Abstract
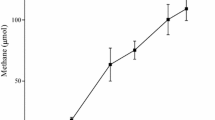
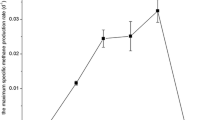
Microbial assemblage in an n-alkanes-dependent thermophilic methanogenic enrichment cultures derived from production waters of a high-temperature petroleum reservoir was investigated in this study. Substantially higher amounts of methane were generated from the enrichment cultures incubated at 55 °C for 528 days with a mixture of long-chain n-alkanes (C15–C20). Stoichiometric estimation showed that alkanes-dependent methanogenesis accounted for about 19.8% of the total amount of methane expected. Hydrogen was occasionally detected together with methane in the gas phase of the cultures. Chemical analysis of the liquid cultures resulted only in low concentrations of acetate and formate. Phylogenetic analysis of the enrichment revealed the presence of several bacterial taxa related to Firmicutes, Thermodesulfobiaceae, Thermotogaceae, Nitrospiraceae, Dictyoglomaceae, Candidate division OP8 and others without close cultured representatives, and Archaea predominantly related to uncultured members in the order Archaeoglobales and CO2-reducing methanogens. Screening of genomic DNA retrieved from the alkanes-amended enrichment cultures also suggested the presence of new alkylsuccinate synthase alpha-subunit (assA) homologues. These findings suggest the presence of poorly characterized (putative) anaerobic n-alkanes degraders in the thermophilic methanogenic enrichment cultures. Our results indicate that methanogenesis of alkanes under thermophilic condition is likely to proceed via syntrophic acetate and/or formate oxidation linked with hydrogenotrophic methanogenesis.





Similar content being viewed by others
References
Aitken CM, Jones DM, Larter SR (2004) Anaerobic hydrocarbon biodegradation in deep subsurface oil reservoirs. Nature 431:291–294
Altschul S, Madden T, Schaffer A, Zhang J, Zhang Z, Miller W, Lipman D (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ (2006) New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl Environ Microbiol 72:5734–5741
Beeder J, Nilsen RK, Rosnes JT, Torsvik T, Lien T (1994) Archaeoglobus fulgidus isolated from hot North Sea oil field waters. Appl Environ Microbiol 60:1227–1231
Beller HR, Kane SR, Legler TC, McKelvie JR, Sherwood Lollar B, Pearson F, Balser L, Mackay DM (2008) Comparative assessments of benzene, toluene, and xylene natural attenuation by quantitative polymerase chain reaction analysis of a catabolic gene, signature metabolites, and compound-specific isotope analysis. Environ Sci Technol 42:6065–6072
Bonin A, Boone D (2006) The order Methanobacteriales. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (eds) The prokaryotes: archaea. Bacteria: Firmicutes, Actinomycetes, 3rd edn. Springer, New York, pp 231–243
Bryant MP (1972) Commentary on the Hungate technique for culture of anaerobic bacteria. Am J Clin Nutr 25:1324–1328
Callaghan AV, Davidova IA, Savage-Ashlock K, Parisi VA, Gieg LM, Suflita JM, Kukor JJ, Wawrik B (2010) Diversity of benzyl- and alkylsuccinate synthase genes in hydrocarbon-impacted environments and enrichment cultures. Environ Sci Technol 44:7287–7294
Chen C-L, Macarie H, Ramirez I, Olmos A, Ong SL, Monroy O, Liu W-T (2004) Microbial community structure in a thermophilic anaerobic hybrid reactor degrading terephthalate. Microbiology 150:3429–3440
Cheng L, Tian-Lei Q, Li X, Wang W-D, Deng Y, Yin X-B, Zhang H (2008) Isolation and characterization of Methanoculleus receptaculi sp. nov. from Shengli oil field, China. FEMS Microbiol Lett 285:65–71
Cheng L, Dai L, Li X, Zhang H, Lu Y (2011) Isolation and characterization of Methanothermobacter crinale sp. nov., a novel hydrogenotrophic methanogen from the Shengli oil field. Appl Environ Microbiol 77:5212–5219
Dojka MA, Hugenholtz P, Haack SK, Pace NR (1998) Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl Environ Microbiol 64:3869–3877
Duncan KE, Gieg LM, Parisi VA, Tanner RS, Tringe SG, Bristow J, Suflita JM (2009) Biocorrosive thermophilic microbial communities in Alaskan North Slope oil facilities. Environ Sci Technol 43:7977–7984
Etchebehere C, Pavan ME, Zorzopulos J, Soubes M, Muxi L (1998) Coprothermobacter platensis sp. nov., a new anaerobic proteolytic thermophilic bacterium isolated from an anaerobic mesophilic sludge. Int J Syst Bacteriol 48:1297–1304
Fang HHP, Liang DW, Zhang T, Liu Y (2006) Anaerobic treatment of phenol in wastewater under thermophilic condition. Water Res 40:427–434
Fardeau M-L, Ollivier B, Patel BKC, Magot M, Thomas P, Rimbault A, Rocchiccioli F, Garcia J-L (1997) Thermotoga hypogea sp. nov., a xylanolytic, thermophilic bacterium from an oil-producing well. Int J Syst Bacteriol 47:1013–1019
Garcia J-L, Ollivier B, Whitman W (2006) The order Methanomicrobiales. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (eds) The prokaryotes: archaea. Bacteria: Firmicutes, Actinomycetes, 3rd edn. Springer, New York, pp 208–230
Gieg LM, Suflita JM (2002) Detection of anaerobic metabolites of saturated and aromatic hydrocarbons in petroleum-contaminated aquifers. Environ Sci Technol 36:3755–3762
Gieg LM, Davidova IA, Duncan KE, Suflita JM (2010) Methanogenesis, sulfate reduction and crude oil biodegradation in hot Alaskan oilfields. Environ Microbiol 12:3074–3086
Good IJ (1953) The population frequencies of species and the estimation of population parameters. Biometrika 40:237–264
Gray ND, Sherry A, Hubert C, Dolfing J, Head IM (2010) Methanogenic degradation of petroleum hydrocarbons in subsurface environments: remediation, heavy oil formation, and energy recovery. Adv Appl Microbiol 72:137–161
Gray ND, Sherry A, Grant RJ, Rowan AK, Hubert CRJ, Callbeck CM, Aitken CM, Jones DM, Adams JJ, Larter SR, Head IM (2011) The quantitative significance of Syntrophaceae and syntrophic partnerships in methanogenic degradation of crude oil alkanes. Environ Microbiol 13:2957–2975
Hales B, Edwards C, Ritchie D, Hall G, Pickup R, Saunders J (1996) Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification and sequence analysis. Appl Environ Microbiol 62:668–675
Head IM, Jones DM, Larter SR (2003) Biological activity in the deep subsurface and the origin of heavy oil. Nature 426:344–352
Holmes DE, Risso C, Smith JA, Lovley DR (2011) Anaerobic oxidation of benzene by the hyperthermophilic archaeon Ferroglobus placidus. Appl Environ Microbiol 77:5926–5933
Huber T, Faulkner G, Hugenholtz P (2004) Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319
Hugenholtz P, Pitulle C, Hershberger KL, Pace NR (1998) Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol 180:366–376
Jones DM, Head IM, Gray ND, Adams JJ, Rowan AK, Aitken CM, Bennett B, Huang H, Brown A, Bowler BFJ, Oldenburg T, Erdmann M, Larter SR (2008) Crude-oil biodegradation via methanogenesis in subsurface petroleum reservoirs. Nature 451:176–180
Khelifi N, Grossi V, Hamdi M, Dolla A, Tholozan J-L, Ollivier B, Hirschler-Rea A (2010) Anaerobic oxidation of fatty acids and alkenes by the hyperthermophilic sulfate-reducing archaeon Archaeoglobus fulgidus. Appl Environ Microbiol 76:3057–3060
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Li H, Yang S-Z, Mu B-Z (2007a) Phylogenetic diversity of the archaeal community in a continental high-temperature, water-flooded petroleum reservoir. Curr Microbiol 55:382–388
Li H, Yang S-Z, Mu B-Z, Rong Z-F, Zhang J (2007b) Molecular phylogenetic diversity of the microbial community associated with a high-temperature petroleum reservoir at an offshore oilfield. FEMS Microbiol Ecol 60:74–84
Li H, Chen S, Mu B-Z, Gu J-D (2010) Molecular detection of anaerobic ammonium-oxidizing (anammox) bacteria in high-temperature petroleum reservoirs. Microb Ecol 60:771–783
Li H, Mu B-Z, Jiang Y, Gu J-D (2011) Production processes affected prokaryotic amoA gene abundance and distribution in high-temperature petroleum reservoirs. Geomicrobiol J 28:692–704
Magot M, Ollivier B, Patel BKC (2000) Microbiology of petroleum reservoirs. Antonie Leeuwenhoek 77:103–116
Mayumi D, Mochimaru H, Yoshioka H, Sakata S, Maeda H, Miyagawa Y, Ikarashi M, Takeuchi M, Kamagata Y (2011) Evidence for syntrophic acetate oxidation coupled to hydrogenotrophic methanogenesis in the high-temperature petroleum reservoir of Yabase oil field (Japan). Environ Microbiol 13:1995–2006
Mbadinga SM, Wang L-Y, Zhou L, Liu J-F, Gu J-D, Mu B-Z (2011) Microbial communities involved in anaerobic degradation of alkanes. Int Biodeterior Biodegrad 65:1–13
McGinnis S, Madden TL (2004) BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res 32:W20–W25
Mochimaru H, Yoshioka H, Tamaki H, Nakamura K, Kaneko N, Sakata S, Imachi H, Sekiguchi Y, Uchiyama H, Kamagata Y (2007) Microbial diversity and methanogenic potential in a high temperature natural gas field in Japan. Extremophiles 11:453–461
Mori K, Kim H, Kakegawa T, Hanada S (2003) A novel lineage of sulfate-reducing microorganisms: Thermodesulfobiaceae fam. nov., Thermodesulfobium narugense, gen. nov., sp. nov., a new thermophilic isolate from a hot spring. Extremophiles 7:283–290
Nazina T, Shestakova N, Grigor’yan A, Mikhailova E, Tourova T, Poltaraus A, Feng C, Ni F, Belyaev S (2006) Phylogenetic diversity and activity of anaerobic microorganisms of high-temperature horizons of the Dagang oil field (P. R. China). Microbiology 75:55–65
Nielsen MB, Kjeldsen KU, Ingvorsen K (2006) Desulfitibacter alkalitolerans gen. nov., sp. nov., an anaerobic, alkalitolerant, sulfite-reducing bacterium isolated from a district heating plant. Int J Syst Evol Microbiol 56:2831–2836
Pouliot J, Galand PE, Lovejoy C, Vincent WF (2009) Vertical structure of archaeal communities and the distribution of ammonia monooxygenase A gene variants in two meromictic High Arctic lakes. Environ Microbiol 11:687–699
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196
Rabus R, Wilkes H, Behrends A, Armstroff A, Fischer T, Pierik AJ, Widdel F (2001) Anaerobic initial reaction of n-alkanes in a denitrifying bacterium: evidence for (1-methylpentyl)succinate as initial product and for involvement of an organic radical in n-hexane metabolism. J Bacteriol 183:1707–1715
Rabus R, Jarling R, Lahme S, Kühner S, Heider J, Widdel F, Wilkes H (2011) Co-metabolic conversion of toluene in anaerobic n-alkane-degrading bacteria. Environ Microbiol 13:2576–2586
Robertson CE, Spear JR, Harris JK, Pace NR (2009) Diversity and stratification of archaea in a hypersaline microbial mat. Appl Environ Microbiol 75:1801–1810
Saiki T, Kobayashi Y, Kawagoe K, Beppu T (1985) Dictyoglomus thermophilum gen. nov., sp. nov., a chemoorganotrophic, anaerobic, thermophilic bacterium. Int J Syst Bacteriol 35:253–259
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Savage KN, Krumholz LR, Gieg LM, Parisi VA, Suflita JM, Allen J, Philp RP, Elshahed MS (2010) Biodegradation of low-molecular-weight alkanes under mesophilic, sulfate-reducing conditions: metabolic intermediates and community patterns. FEMS Microbiol Ecol 72:485–495
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Siddique T, Penner T, Semple K, Foght JM (2011) Anaerobic biodegradation of longer-chain n-alkanes coupled to methane production in oil sands tailings. Environ Sci Technol 45:5892–5899
Spear JR, Walker JJ, McCollom TM, Pace NR (2005) Hydrogen and bioenergetics in the Yellowstone geothermal ecosystem. Proc Natl Acad Sci USA 102:2555–2560
Stetter KO, Lauerer G, Thomm M, Neuner A (1987) Isolation of extremely thermophilic sulfate reducers: evidence for a novel branch of Archaebacteria. Science 236:822–824
Symons GE, Buswell AM (1933) The methane fermentation of carbohydrates1,2. J Am Chem Soc 55:2028–2036
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Vasconcellos SPd, Angolini CFF, García INS, Martins Dellagnezze B, Silva CCd, Marsaioli AJ, Neto EVdS, de Oliveira VM (2010) Screening for hydrocarbon biodegraders in a metagenomic clone library derived from Brazilian petroleum reservoirs. Org Geochem 41:675–681
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Wang L-Y, Gao C-X, Mbadinga SM, Zhou L, Liu J-F, Gu J-D, Mu B-Z (2011) Characterization of an alkane-degrading methanogenic enrichment culture from production water of an oil reservoir after 274 days of incubation. Int Biodeterior Biodegrad 65:444–450
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Widdel F, Grundmann O (2010) Biochemistry of the anaerobic degradation of non-methane alkanes. In: Timmis KN (ed) Handbook of hydrocarbon and lipid microbiology. Springer, Berlin, pp 909–924
Wrighton KC, Agbo P, Warnecke F, Weber KA, Brodie EL, DeSantis TZ, Hugenholtz P, Andersen GL, Coates JD (2008) A novel ecological role of the Firmicutes identified in thermophilic microbial fuel cells. ISME J 2:1146–1156
Yu Y, Breitbart M, McNairnie P, Rohwer F (2006) FastGroupII: a web-based bioinformatics platform for analyses of large 16S rDNA libraries. BMC Bioinforma 7:57
Zengler K, Richnow HH, Rossello-Mora R, Michaelis W, Widdel F (1999) Methane formation from long-chain alkanes by anaerobic microorganisms. Nature 401:266–269
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant no. 51174092) and the 863 Program (grant no. 2009AA063503).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary materials
Below is the link to the electronic supplementary material.
ESM 1
(PDF 147 kb)
Rights and permissions
About this article
Cite this article
Mbadinga, S.M., Li, KP., Zhou, L. et al. Analysis of alkane-dependent methanogenic community derived from production water of a high-temperature petroleum reservoir. Appl Microbiol Biotechnol 96, 531–542 (2012). https://doi.org/10.1007/s00253-011-3828-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3828-8