Abstract
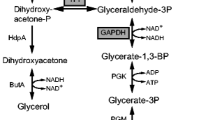
We previously demonstrated the simplicity of oxygen-deprived Corynebacterium glutamicum to produce d-lactate, a primary building block of next-generation biodegradable plastics, at very high optical purity by introducing heterologous D-ldhA gene from Lactobacillus delbrueckii. Here, we independently evaluated the effects of overexpressing each of genes encoding the ten glycolytic enzymes on d-lactate production in C. glutamicum. We consequently show that while the reactions catalyzed by glucokinase (GLK), glyceraldehyde 3-phosphate dehydrogenase (GAPDH), phosphofructokinase (PFK), triosephosphate isomerase (TPI), and bisphosphate aldolase had positive effects on d-lactate productivity by increasing 98, 39, 15, 13, and 10 %, respectively, in 10 h reactions in minimal salts medium, the reaction catalyzed by pyruvate kinase had large negative effect by decreasing 70 %. The other glycolytic enzymes did not affect d-lactate productivity when each of encoding genes was overexpressed. It is noteworthy that all reactions associated with positive effects are located upstream of glycerate-1,3-bisphosphate in the glycolytic pathway. The d-lactate yield also increased by especially overexpressing TPI encoding gene up to 94.5 %. Interestingly, overexpression of PFK encoding gene reduced the yield of succinate, one of the main by-products of d-lactate production, by 52 %, whereas overexpression of GAPDH encoding gene increased succinate yield by 26 %. Overexpression of GLK encoding gene markedly increased the yield of dihydroxyacetone and glycerol by 10- and 5.8-fold in exchange with decreasing the d-lactate yield. The effect of overexpressing glycolytic genes was also evaluated in 80 h long-term reactions. The variety of effects of overexpressing each of genes encoding the ten glycolytic enzymes on d-lactate production is discussed.



Similar content being viewed by others
References
Babul J, Clifton D, Kretschmer M, Fraenkel DG (1993) Glucose metabolism in Escherichia coli and the effect of increased amount of aldolase. Biochemistry 32:4685–4692
Blombach B, Riester T, Wieschalka S, Ziert C, Youn JW, Wendisch VF, Eikmanns BJ (2011) Corynebacterium glutamicum tailored for efficient isobutanol production. Appl Environ Microbiol 77:3300–3310
Davies SE, Brindle KM (1992) Effects of overexpression of phosphofructokinase on glycolysis in the yeast Saccharomyces cerevisiae. Biochemistry 31:4729–4735
Dominguez H, Rollin C, Guyonvarch A, Guerquin-Kern JL, Cocaign-Bousquet M, Lindley ND (1998) Carbon-flux distribution in the central metabolic pathways of Corynebacterium glutamicum during growth on fructose. Eur J Biochem 254:96–102
Emmerling M, Bailey JE, Sauer U (2000) Altered regulation of pyruvate kinase or co-overexpression of phosphofructokinase increases glycolytic fluxes in resting Escherichia coli. Biotechnol Bioeng 67:623–627
Fukushima K, Chang YH, Kimura Y (2007) Enhanced stereocomplex formation of poly(L-lactic acid) and poly (D-lactic acid) in the presence of stereoblock poly(lactic acid). Macromol Biosci 7:829–835
Hasegawa S, Uematsu K, Natsuma Y, Suda M, Hiraga K, Jojima T, Inui M, Yukawa H (2012) Improvement of the redox balance increases L-valine production by Corynebacterium glutamicum under oxygen deprivation conditions. Appl Environ Microbiol 78:865–875
Hatzimanikatis V, Emmerling M, Sauer U, Bailey JE (1998) Application of mathematical tools for metabolic design of microbial ethanol production. Biotechnol Bioeng 58:154–161
Ikada Y, Jamshidi K, Tsuji H, Hyon S (1987) Stereocomplex formation between enantiomeric poly (lactides). Macromolecules 20: 904–906
Ikeda M, Mizuno Y, Awane S, Hayashi M, Mitsuhashi S, Takeno S. (2011) Identification and application of a different glucose uptake system that functions as an alternative to the phosphotransferase system in Corynebacterium glutamicum. Appl Microbiol Biotechnol 90:1443–1451
Inui M, Nakata K, Roh JH, Vertès AA, Yukawa H (2003) Isolation and molecular characterization of pMG160, a mobilizable cryptic plasmid from Rhodobacter blasticus. Appl Environ Microbiol 69:725–733
Inui M, Kawaguchi H, Murakami S, Vertès AA, Yukawa H (2004a) Metabolic engineering of Corynebacterium glutamicum for fuel ethanol production under oxygen-deprivation conditions. J Mol Microbiol Biotechnol 8:243–254
Inui M, Murakami S, Okino S, Kawaguchi H, Vertès AA, Yukawa H (2004b) Metabolic analysis of Corynebacterium glutamicum during lactate and succinate productions under oxygen deprivation conditions. J Mol Microbiol Biotechnol 7:182–196
Inui M, Suda M, Okino S, Nonaka H, Puskas LG, Vertès AA, Yukawa H (2007) Transcriptional profiling of Corynebacterium glutamicum metabolism during organic acid production under oxygen deprivation conditions. Microbiology 153:2491–2504
Jojima T, Fujii M, Mori E, Inui M, Yukawa H (2010) Engineering of sugar metabolism of Corynebacterium glutamicum for production of amino acid L-alanine under oxygen deprivation. Appl Microbiol Biotechnol 87:159–165
Karjomaa S, Suortti T, Lempiainen R, Selin J, Itavaara M (1998) Microbial degradation of poly-(L-lactic acid) oligomers. Polym Degrad Stab 59:333–336
Kinoshita S (1985) Glutamic acid bacteria. In: Demain AL, Solomon NA (ed) Biology of industrial microorganisms. Cummings, London, pp 115–146
Koebmann BJ, Solem C, Pedersen MB, Nilsson D, Jensen PR (2002a) Expression of genes encoding F(1)-ATPase results in uncoupling of glycolysis from biomass production in Lactococcus lactis. Appl Environ Microbiol 68:4274–4282
Koebmann BJ, Westerhoff HV, Snoep JL, Nilsson D, Jensen PR (2002b) The glycolytic flux in Escherichia coli is controlled by the demand for ATP. J Bacteriol 184:3909–3916
Larsson C, Nilsson A, Blomberg A, Gustafsson L. (1997) Glycolytic flux is conditionally correlated with ATP concentration in Saccharomyces cerevisiae: a chemostat study under carbon- or nitrogen-limiting conditions. J Bacteriol 179:7243–7250.
Lindner SN, Seibold GM, Henrich A, Krämer R, Wendisch VF. (2011) Phosphotransferase system-independent glucose utilization in Corynebacterium glutamicum by inositol permeases and glucokinases. Appl Environ Microbiol 77:3571–3581
Litsanov B, Brocker M, Bott M (2012) Toward homosuccinate fermentation: metabolic engineering of Corynebacterium glutamicum for anaerobic production of succinate from glucose and formate. Appl Environ Microbiol 78:3325–3337
Nakayama K, Kitada S, Kinoshita S (1961) Studies on lysine fermentation I. The control mechanism on lysine accumulation by homoserine and threonine. J Gen Appl Microbiol 7:145–154
Okano K, Yoshida S, Yamada R, Tanaka T, Ogino C, Fukuda H, Kondo A (2009) Improved production of homo-D-lactic acid via xylose fermentation by introduction of xylose assimilation genes and redirection of the phosphoketolase pathway to the pentose phosphate pathway in L-Lactate dehydrogenase gene-deficient Lactobacillus plantarum. Appl Environ Microbiol 75:7858–7861
Okino S, Inui M, Yukawa H (2005) Production of organic acids by Corynebacterium glutamicum under oxygen deprivation. Appl Microbiol Biotechnol 68:475–480
Okino S, Noburyu R, Suda M, Jojima T, Inui M, Yukawa H (2008a) An efficient succinic acid production process in a metabolically engineered Corynebacterium glutamicum strain. Appl Microbiol Biotechnol 81:459–464
Okino S, Suda M, Fujikura K, Inui M, Yukawa H (2008b) Production of D-lactic acid by Corynebacterium glutamicum under oxygen deprivation. Appl Microbiol Biotechnol 78:449–454
Papagianni M, Avramidis N (2012) Lactococcus lactis as a cell factory: a twofold increase in phosphofructokinase activity results in a proportional increase in specific rates of glucose uptake and lactate formation. Enzyme Microb Technol 49:197–202
Ramos A, Neves AR, Ventura R, Maycock C, Lopez P, Santos H (2004) Effect of pyruvate kinase overproduction on glucose metabolism of Lactococcus lactis. Microbiology 150:1103–1111
Rud I, Solem C, Jensen PR, Axelsson L, Naterstad K (2008) Co-factor engineering in lactobacilli: effects of uncoupled ATPase activity on metabolic fluxes in Lactobacillus plantarum and L. sakei. Metab Eng 10(5):207–215
Sambrook J, Fritsh E, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Santamaria R, Gil J, Mesas J, Martin J (1984) Characterization of an endogenous plasmid and development of cloning vectors and a transformation system in Brevibacterium lactofermentum. J Gen Microbiol 130:2237–2246
Schaaff I, Heinisch J, Zimmermann FK (1989) Overproduction of glycolytic enzymes in yeast. Yeast 5:285–290
Sekine H, Shimada T, Hayashi C, Ishiguro A, Tomita F, Yokota A (2001) H+-ATPase defect in Corynebacterium glutamicum abolishes glutamic acid production with enhancement of glucose consumption rate. Appl Microbiol Biotechnol 57:534–540
Solem C, Koebmann BJ, Jensen PR (2003) Glyceraldehyde-3-phosphate dehydrogenase has no control over glycolytic flux in Lactococcus lactis MG1363. J Bacteriol 185:1564–1571
Snoep J, Yomano L, Westerhoff H, Ingram L (1995) Protein burden in Zymomonas mobilis: negative flux and growth control due to overproduction of glycolytic enzymes. Microbiology 141:2329–2337
Suzuki N, Okayama S, Nonaka H, Tsuge Y, Inui M, Yukawa H (2005) Large-scale engineering of the Corynebacterium glutamicum genome. Appl Environ Microbiol 71:3369–3372
Tsuchida Y, Kimura S, Suzuki N, Inui M, Yukawa H (2009) Characterization of a 24-kb plasmid pCGR2 newly isolated from Corynebacterium glutamicum. Appl Microbiol Biotechnol 87:1855–1866
Tsuge Y, Suzuki N, Inui M, Yukawa H (2007) Random segment deletion based on IS31831 and Cre/loxP excision system in Corynebacterium glutamicum. Appl Microbiol Biotechnol 74:1333–1341
Utrilla J, Gosset G, Martinez A (2009) ATP limitation in a pyruvate formate lyase mutant of Escherichia coli MG1655 increases glycolytic flux to D-lactate. J Ind Microbiol Biotechnol 36:1057–1062
Wittmann C, Kiefer P, Zelder O (2004) Metabolic fluxes in Corynebacterium glutamicum during lysine production with sucrose as carbon source. Appl Environ Microbiol 70:7277–7287
Yamamoto S, Gunji W, Suzuki H, Toda H, Suda M, Jojima T, Inui M, Yukawa H (2012) Overexpression of glycolytic genes enhances Corynebacterium glutamicum glucose metabolism and alanine production under oxygen-deprived conditions. Appl Environ Microbiol 78:4447–4457
Yoshida S, Okano K, Tanaka T, Ogino C, Kondo A (2011) Homo-D-lactic acid production from mixed sugars using xylose-assimilating operon-integrated Lactobacillus plantarum. Appl Microbiol Biotechnol 92:67–76
Yukawa H, Omumasaba CA, Nonaka H, Kos P, Okai N, Suzuki N, Suda M, Tsuge Y, Watanabe J, Ikeda Y, Vertès AA, Inui M (2007) Comparative analysis of the Corynebacterium glutamicum group and complete genome sequence of strain R. Microbiology 153:1042–1058
Zhou S, Causey TB, Hasona A, Shanmugam KT, Ingram LO (2003) Production of optically pure D-lactic acid in mineral salts medium by metabolically engineered Escherichia coli W3110. Appl Environ Microbiol 69:399–407
Zhu J, Shimizu K (2005) Effect of a single-gene knockout on the metabolic regulation in Escherichia coli for D-lactate production under microaerobic condition. Metab Eng 7:104–115
Acknowledgment
We thank Crispinus A. Omumasaba for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tsuge, Y., Yamamoto, S., Suda, M. et al. Reactions upstream of glycerate-1,3-bisphosphate drive Corynebacterium glutamicum d-lactate productivity under oxygen deprivation. Appl Microbiol Biotechnol 97, 6693–6703 (2013). https://doi.org/10.1007/s00253-013-4986-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-013-4986-7