Abstract
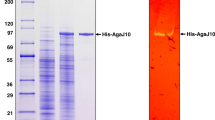
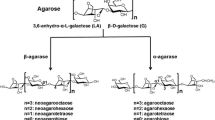
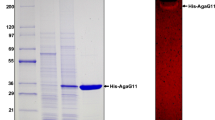
An agar-degrading bacterium, Catenovulum sp. X3, was isolated from the seawater of Shantou, China. A novel β-agarase gene agaXa was cloned from the strain Catenovulum sp. X3. The gene agaXa consists of 1,590 bp and encodes a protein of 529 amino acids, with only 40 % amino acid sequence identity with known agarases. AgaXa should belong to the glycoside hydrolase family GH118 based on the amino acid sequence similarity. The molecular mass of the recombinant AgaXa (rAgaXa) was estimated to be 52 kDa by sodium dodecyl sulfate–polyacrylamide gel electrophoresis. It had a maximal agarase activity at 52 °C and pH 7.4 and was stable over pH 5.0 ~ 9.0 and at temperatures below 42 °C. The K m and V max for agarose were 10.5 mg/ml and 588.2 U/mg, respectively. The purified rAgaXa showed endolytic activity on agarose degradation, yielding neoagarohexaose, neoagarooctaose, neoagarodecaose, and neoagarododecaose as the end products. The results showed that AgaXa has potential applications in agar degradation for the production of oligosaccharides with various bioactivities.






Similar content being viewed by others
References
Bannikova GE, Lopatin SA, Varlamov VP, Kuznetsov BB, Kozina IV, Miroshnichenko ML, Chernykh NA, Turova TP, Bonch-Osmolovskaya EA (2008) The thermophilic bacteria hydrolyzing agar: characterization of thermostable agarase. Appl Biochem Microbiol 45:366–371
Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B (2009) The Carbohydrate-Active EnZyme database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:D233–238
Chen LCM, Craigiel JS, Xie ZK (1994) Protoplast production from Porphyra linearis using a simplified agarase procedure capable of commercial application. J Appl Phycol 6:35–39
Chi WJ, Chang YK, Hong SK (2012) Agar degradation by microorganisms and agar-degrading enzymes. Appl Microbiol Biotechnol 94:917–930
Dipakkore S, Reddy CRK, Jha B (2005) Production and seeding of protoplasts of Porphyra okhaensis (Bangiales, Rhodophyta) in laboratory culture. J Appl Phycol 17:331–337
Dong J, Hashikawa S, Konishi T, Tamaru Y, Araki T (2006) Cloning of the novel gene-encoding beta-agarase C from a marine bacterium, Vibrio sp. strain PO-303, and characterization of the gene product. Appl Environ Microbiol 72:6399–6401
Duckworth M, Yaphe W (1971) Structure of agar: part I. Fractionation of a complex mixture of polysaccharides. Carbohydr Res 16:189–197
Giordano A, Andreotti G, Tramice A, Trincone A (2006) Marine glycosyl hydrolases in the hydrolysis and synthesis of oligosaccharides. J Biotechnol 1:511–530
Finkelstein M, Rownd RH (1978) A rapid method for extracting DNA from agarose gels. Plasmid 1:557–562
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kobayashi R, Takimasa M, Suzuki T, Kirimura K, Usami S (1997) Neoagarobiose as a novel moisturizer with whitening effect. Biosci Biotechnol Biochem 61:162–163
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Ltd., Chichester, pp 115–175
Lee DG, Jang MK, Lee OH, Kim NY, Ju SA, Lee SH (2008) Overproduction of a glycoside hydrolase family 50 beta-agarase from Agarivorans sp. JA-1 in Bacillus subtilis and the whitening effect of its product. Biotechnol Lett 30:911–918
Lin B, Lu G, Zheng Y, Xie W, Li S, Hu Z (2012a) Aquimarina agarilytica sp. nov., a novel agarolytic species isolated from red alga. Int J Syst Evol Microbiol 62:869–873
Lin B, Lu G, Zheng Y, Xie W, Li S, Hu Z (2012b) Gene cloning, expression and characterization of a neoagarotetraose-producing β-agarase from a marine bacterium Agarivorans sp. HZ105. World J Microb Biot 28:1691–1697
Ma CP, Lu XZ, Shi C, Li J, Gu Y, Ma Y, Chu Y, Han F, Gong Q, Yu W (2007) Molecular cloning and characterization of a novel β-agarase, AgaB, from marine Pseudoalteromonas sp. CY24. J Biol Chem 282:3747–3754
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428
Ohta Y, Hatada Y, Nogi Y, Miyazaki M, Li Z, Akita M, Hidaka Y, Goda S, Ito S, Horikoshi K (2004a) Enzymatic properties and nucleotide and amino acid sequences of a thermostable β-agarase from a novel species of deep-sea microbulbifer. Appl Microbiol Biotechnol 64:505–514
Ohta Y, Nogi Y, Miyazaki M, Li Z, Hatada Y, Ito S, Horikoshi K (2004b) Enzymatic properties and nucleotide and amino acid sequences of a thermostable β-agarase from the novel marine isolate JAMB-A94. Biosci Biotechnol Biochem 68:1073–1081
Oppenheimer CH, ZoBell CE (1952) The growth and viability of sixty-three species of marine bacteria as influenced by hydrostatic pressure. J Mar Res 11:10–18
Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786
Ren A, Xia ZX, Yu W, Zhou J (2010) Expression, crystallization and preliminary X-ray analysis of an anomeric inverting agarase from Pseudoalteromonas sp. CY24. Acta Cryst F66:1635–1639
Wu SC, Wen TN, Pan CL (2005) Algal–oligosaccharide–lysates prepared by two bacterial agarases stepwise hydrolyzed and their antioxidative properties. Fish Sci 71:1149–1159
Yan S, Yu M, Wang Y, Shen C, Zhang XH (2011) Catenovulum agarivorans gen. nov., sp. nov., a peritrichously flagellated, chain-forming, agar-hydrolysing gammaproteobacterium from seawater. Int J Syst Evol Microbiol 61:2866–2873
Zhang WW, Sun L (2007) Cloning, characterization, and molecular application of a beta-agarase gene from Vibrio sp. strain V134. Appl Environ Microbiol 73:2825–2831
Acknowledgements
This work was supported by the National Natural Science Foundation of China (nos. 41076106 and 31200077), Guangdong Natural Science Foundation (no. S2011030005257), and the Key Science and Technology Innovation Project for University by the Department of Education of Guangdong Province (no. CXZD1124) and the Science & Technology Project of Guangdong Province (no. 2012A031100009).
Author information
Authors and Affiliations
Corresponding author
Additional information
Wei Xie, Bokun Lin, and Zhengrong Zhou contributed equally to this work.
Rights and permissions
About this article
Cite this article
Xie, W., Lin, B., Zhou, Z. et al. Characterization of a novel β-agarase from an agar-degrading bacterium Catenovulum sp. X3. Appl Microbiol Biotechnol 97, 4907–4915 (2013). https://doi.org/10.1007/s00253-012-4385-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-012-4385-5