Abstract
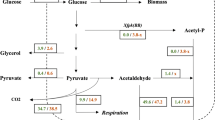
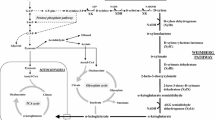
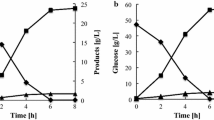
Three enzymes responsible for the transhydrogenase-like shunt, including malic enzyme (encoded by MAE1), malate dehydrogenase (MDH2), and pyruvate carboxylase (PYC2), were overexpressed to regulate the redox state in xylose-fermenting recombinant Saccharomyces cerevisiae. The YPH499XU/MAE1 strain was constructed by overexpressing native Mae1p in the YPH499XU strain expressing xylose reductase and xylitol dehydrogenase from Scheffersomyces stipitis, and native xylulokinase. Analysis of the xylose fermentation profile under semi-anaerobic conditions revealed that the ethanol yield in the YPH499XU/MAE1 strain (0.38 ± 0.01 g g−1 xylose consumed) was improved from that of the control strain (0.31 ± 0.01 g g−1 xylose consumed). Reduced xylitol production was also observed in YPH499XU/MAE1, suggesting that the redox balance was altered by Mae1p overexpression. Analysis of intracellular metabolites showed that the redox imbalance during xylose fermentation was partly relieved in the transformant. The specific ethanol production rate in the YPH499XU/MAE1–MDH2 strain was 1.25-fold higher than that of YPH499XU/MAE1 due to the additional overexpression of Mdh2p, whereas the ethanol yield was identical to that of YPH499XU/MAE1. The specific xylose consumption rate was drastically increased in the YPH499XU/MAE1–MDH2–PYC2 strain. However, poor ethanol yield as well as increased production of xylitol was observed. These results demonstrate that the transhydrogenase function implemented in S. cerevisiae can regulate the redox state of yeast cells.




Similar content being viewed by others
References
Amore R, Kotter P, Kuster C, Ciriacy M, Hollenberg CP (1991) Cloning and expression in Saccharomyces cerevisiae of the NAD(P)H-dependent xylose reductase-encoding gene (XYL1) from the xylose-assimilating yeast Pichia stipitis. Gene 109:89–97
Anderlund M, Nissen TL, Nielsen J, Villadsen J, Rydstrom J, Hahn-Hagerdal B, Kielland-Brandt MC (1999) Expression of the Escherichia coli pntA and pntB genes, encoding nicotinamide nucleotide transhydrogenase, in Saccharomyces cerevisiae and its effect on product formation during anaerobic glucose fermentation. Appl Environ Microbiol 65:2333–2340
Arkblad EL, Betsholtz C, Rydstrom J (1996) The cDNA sequence of proton-pumping nicotinamide nucleotide transhydrogenase from man and mouse. Biochim Biophys Acta 1273:203–205
Bakker BM, Overkamp KM, van Maris AJ, Kotter P, Luttik MA, van Dijken JP, Pronk JT (2001) Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol Rev 25:15–37
Blombach B, Riester T, Wieschalka S, Ziert C, Youn JW, Wendisch VF, Eikmanns BJ (2011) Corynebacterium glutamicum tailored for efficient isobutanol production. Appl Environ Microbiol 77:3300–3310
Boles E, de Jong-Gubbels P, Pronk JT (1998) Identification and characterization of MAE1, the Saccharomyces cerevisiae structural gene encoding mitochondrial malic enzyme. J Bacteriol 180:2875–2882
Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115–132
Bro C, Regenberg B, Forster J, Nielsen J (2006) In silico aided metabolic engineering of Saccharomyces cerevisiae for improved bioethanol production. Metab Eng 8:102–111
Bruinenberg PM, de Bot PHM, van Dijken JP, Scheffers WA (1983) The role of redox balances in the anaerobic fermentation of xylose by yeasts. Eur J Appl Microbiol Biotechnol 18:287–292
Eliasson A, Christensson C, Wahlbom CF, Hahn-Hagerdal B (2000) Anaerobic xylose fermentation by recombinant Saccharomyces cerevisiae carrying XYL1, XYL2, and XKS1 in mineral medium chemostat cultures. Appl Environ Microbiol 66:3381–3386
Grotkjaer T, Christakopoulos P, Nielsen J, Olsson L (2005) Comparative metabolic network analysis of two xylose fermenting recombinant Saccharomyces cerevisiae strains. Metab Eng 7:437–444
Hahn-Hagerdal B, Karhumaa K, Jeppsson M, Gorwa-Grauslund MF (2007) Metabolic engineering for pentose utilization in Saccharomyces cerevisiae. Adv Biochem Eng Biotechnol 108:147–177
Hasunuma T, Sung KM, Sanda T, Yoshimura K, Matsuda F, Kondo A (2011) Efficient fermentation of xylose to ethanol at high formic acid concentrations by metabolically engineered Saccharomyces cerevisiae. Appl Microbiol Biotechnol 90:997–1004
Ho NW, Chen Z, Brainard AP (1998) Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl Environ Microbiol 64:1852–1859
Hsiao H-Y, Chiang L-C, Ueng PP, Tsao GT (1982) Sequential utilization of mixed monosaccharides by yeasts. Appl Environ Microbiol 43:840–845
Ishii J, Izawa K, Matsumura S, Wakamura K, Tanino T, Tanaka T, Ogino C, Fukuda H, Kondo A (2009) A simple and immediate method for simultaneously evaluating expression level and plasmid maintenance in yeast. J Biochem 145:701–708
Ito H, Fukuda Y, Murata K, Kimura A (1983) Transformation of intact yeast cells treated with alkali cations. J Bacteriol 153:163–168
Jeffries TW, Jin YS (2004) Metabolic engineering for improved fermentation of pentoses by yeasts. Appl Microbiol Biotechnol 63:495–509
Jeppsson M, Johansson B, Hahn-Hagerdal B, Gorwa-Grauslund MF (2002) Reduced oxidative pentose phosphate pathway flux in recombinant xylose-utilizing Saccharomyces cerevisiae strains improves the ethanol yield from xylose. Appl Environ Microbiol 68:1604–1609
Jeppsson M, Johansson B, Jensen PR, Hahn-Hagerdal B, Gorwa-Grauslund MF (2003) The level of glucose-6-phosphate dehydrogenase activity strongly influences xylose fermentation and inhibitor sensitivity in recombinant Saccharomyces cerevisiae strains. Yeast 20:1263–1272
Johansson B, Hahn-Hagerdal B (2002) The non-oxidative pentose phosphate pathway controls the fermentation rate of xylulose but not of xylose in Saccharomyces cerevisiae TMB3001. FEMS Yeast Res 2:277–282
Kötter P, Ciriacy M (1993) Xylose fermentation by Saccharomyces cerevisiae. Appl Microbiol Biotechnol 38:776–783
Katahira S, Ito M, Takema H, Fujita Y, Tanino T, Tanaka T, Fukuda H, Kondo A (2008) Improvement of ethanol productivity during xylose and glucose co-fermentation by xylose-assimilating S. cerevisiae via expression of glucose transporter Sut1. Enzyme Microb Technol 43:115–119
Katahira S, Mizuike A, Fukuda H, Kondo A (2006) Ethanol fermentation from lignocellulosic hydrolysate by a recombinant xylose- and cellooligosaccharide-assimilating yeast strain. Appl Microbiol Biotechnol 72:1136–1143
Kato H, Izumi Y, Hasunuma T, Matsuda F, Kondo A (2012) Widely targeted metabolic profiling analysis of yeast central metabolites. J Biosci Bioeng 113:665–673
Klimacek M, Krahulec S, Sauer U, Nidetzky B (2010) Limitations in xylose-fermenting Saccharomyces cerevisiae, made evident through comprehensive metabolite profiling and thermodynamic analysis. Appl Environ Microbiol 76:7566–7574
Krahulec S, Petschacher B, Wallner M, Longus K, Klimacek M, Nidetzky B (2010) Fermentation of mixed glucose–xylose substrates by engineered strains of Saccharomyces cerevisiae: role of the coenzyme specificity of xylose reductase, and effect of glucose on xylose utilization. Microb Cell Fact 9:16
Matsuda F, Furusawa C, Kondo T, Ishii J, Shimizu H, Kondo A (2011) Engineering strategy of yeast metabolism for higher alcohol production. Microb Cell Fact 10:70
Matsushika A, Inoue H, Kodaki T, Sawayama S (2009) Ethanol production from xylose in engineered Saccharomyces cerevisiae strains: current state and perspectives. Appl Microbiol Biotechnol 84:37–53
Matsushika A, Watanabe S, Kodaki T, Makino K, Sawayama S (2008) Bioethanol production from xylose by recombinant Saccharomyces cerevisiae expressing xylose reductase, NADP(+)-dependent xylitol dehydrogenase, and xylulokinase. J Biosci Bioeng 105:296–299
Moreira dos Santos M, Raghevendran V, Kotter P, Olsson L, Nielsen J (2004) Manipulation of malic enzyme in Saccharomyces cerevisiae for increasing NADPH production capacity aerobically in different cellular compartments. Metab Eng 6:352–363
Nevoigt E (2008) Progress in metabolic engineering of Saccharomyces cerevisiae. Microbiol Mol Biol Rev 72:379–412
Nissen TL, Anderlund M, Nielsen J, Villadsen J, Kielland-Brandt MC (2001) Expression of a cytoplasmic transhydrogenase in Saccharomyces cerevisiae results in formation of 2-oxoglutarate due to depletion of the NADPH pool. Yeast 18:19–32
Pitkanen JP, Aristidou A, Salusjarvi L, Ruohonen L, Penttila M (2003) Metabolic flux analysis of xylose metabolism in recombinant Saccharomyces cerevisiae using continuous culture. Metab Eng 5:16–31
Rizzi M, Erlemann P, Bui-Thanh N, Dellweg H (1988) Xylose fermentation by yeasts: 4. Purification and kinetic studies of xylose reductase from Pichia stipitis. Appl Microbiol Biotechnol 29:148–154
Rizzi M, Harwart K, Erlemann P, Bui-Thanh N, Dellweg H (1989) Purification and properties of the NAD + -xylitol-dehydrogenase from the yeast Pichia stipitis. J Ferment Bioeng 67:20–24
Sauer U, Canonaco F, Heri S, Perrenoud A, Fischer E (2004) The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J Biol Chem 279:6613–6619
Sonderegger M, Jeppsson M, Hahn-Hagerdal B, Sauer U (2004) Molecular basis for anaerobic growth of Saccharomyces cerevisiae on xylose, investigated by global gene expression and metabolic flux analysis. Appl Environ Microbiol 70:2307–2317
Sonderegger M, Sauer U (2003) Evolutionary engineering of Saccharomyces cerevisiae for anaerobic growth on xylose. Appl Environ Microbiol 69:1990–1998
Spura J, Reimer LC, Wieloch P, Schreiber K, Buchinger S, Schomburg D (2009) A method for enzyme quenching in microbial metabolome analysis successfully applied to gram-positive and gram-negative bacteria and yeast. Anal Biochem 394:192–201
Tantirungkij M, Nakashima N, Seki T, Yoshida T (1993) Construction of xylose-assimilating Saccharomyces cerevisiae. J Ferment Bioeng :83–88
Van Vleet JH, Jeffries TW (2009) Yeast metabolic engineering for hemicellulosic ethanol production. Curr Opin Biotechnol 20:300–306
Verho R, Londesborough J, Penttila M, Richard P (2003) Engineering redox cofactor regeneration for improved pentose fermentation in Saccharomyces cerevisiae. Appl Environ Microbiol 69:5892–5897
Watanabe S, Abu Saleh A, Pack SP, Annaluru N, Kodaki T, Makino K (2007) Ethanol production from xylose by recombinant Saccharomyces cerevisiae expressing protein-engineered NADH-preferring xylose reductase from Pichia stipitis. Microbiology 153:3044–3054
Acknowledgements
This work was financed by the Industrial Technology Research Grant Program in 2011 of the New Energy and Industrial Technology Development Organization (NEDO), Japan, and Special Coordination Funds for Promoting Science and Technology, Creation of Innovation Centers for Advanced Interdisciplinary Research Areas (Innovative Bioproduction Kobe), MEXT, Japan.
Conflict of interests
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Suga, H., Matsuda, F., Hasunuma, T. et al. Implementation of a transhydrogenase-like shunt to counter redox imbalance during xylose fermentation in Saccharomyces cerevisiae . Appl Microbiol Biotechnol 97, 1669–1678 (2013). https://doi.org/10.1007/s00253-012-4298-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-012-4298-3