Abstract
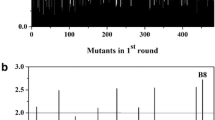
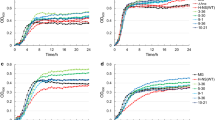
One major challenge in biofuel production, including biobutanol production, is the low tolerance of the microbial host towards increasing biofuel concentration during fermentation. Here, we have demonstrated that Escherichia coli 1-butanol tolerance can be greatly enhanced through random mutagenesis of global transcription factor cyclic AMP receptor protein (CRP). Four mutants (MT1–MT4) with elevated 1-butanol tolerance were isolated from error-prone PCR libraries through an enrichment screening. A DNA shuffling library was then constructed using MT1–MT4 as templates and one mutant (MT5) that exhibited the best tolerance ability among all variants was selected. In the presence of 0.8 % (v/v, 6.5 g/l) 1-butanol, the growth rate of MT5 was found to be 0.28 h−1 while that of wild type was 0.20 h−1. When 1-butanol concentration increased to 1.2 % (9.7 g/l), the growth rate of MT5 (0.18 h−1) became twice that of the wild type (0.09 h−1). Microbial adhesion to hydrocarbon test showed that cell surface of MT5 was less hydrophobic and its cell length became significantly longer in the presence of 1-butanol, as observed by scanning electron microscopy. Quantitative real-time reverse transcription PCR analysis revealed that several CRP regulated, 1-butanol stress response related genes (rpoH, ompF, sodA, manX, male, and marA) demonstrated differential expression in MT5 in the presence or absence of 1-butanol. In conclusion, direct manipulation of the transcript profile through engineering global transcription factor CRP can provide a useful tool in strain engineering.






Similar content being viewed by others
References
Alper H, Stephanopoulos G (2007) Global transcription machinery engineering: a new approach for improving cellular phenotype. Metab Eng 9(3):258–267
Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G (2006) Engineering yeast transcription machinery for improved ethanol tolerance and production. Science 314(5805):1565–1568
Aono R (1998) Improvement of organic solvent tolerance level of Escherichia coli by overexpression of stress-responsive genes. Extremophiles 2(3):239–248
Atsumi S, Liao JC (2008) Metabolic engineering for advanced biofuels production from Escherichia coli. Curr Opin Biotechnol 19(5):414–419
Atsumi S, Hanai T, Liao JC (2008) Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451(7174):86–89, U13
Baker CH, Tomlinson SR, Garcia AE, Harman JG (2001) Amino acid substitution at position 99 affects the rate of CRP subunit exchange. Biochemistry 40(41):12329–12338
Bond-Watts BB, Bellerose RJ, Chang MCY (2011) Enzyme mechanism as a kinetic control element for designing synthetic biofuel pathways. Nat Chem Biol 7(4):222–227
Botsford JL, Harman JG (1992) Cyclic-Amp in Prokaryotes. Microbiol Rev 56(1):100–122
Cascone R (2008) Biobutanol—a replacement for bioethanol? Chem Eng Prog 104(8):S4–S9
Feingersch R, Shainsky J, Wood TK, Fishman A (2008) Protein engineering of toluene monooxygenases for synthesis of chiral sulfoxides. Appl Environ Microbiol 74(5):1555–1566
Fischer CR, Klein-Marcuschamer D, Stephanopoulos G (2008) Selection and optimization of microbial hosts for biofuels production. Metab Eng 10(6):295–304
Gama-Castro S, Salgado H, Peralta-Gil M, Santos-Zavaleta A, Muñiz-Rascado L, Solano-Lira H, Jimenez-Jacinto V, Weiss V, García-Sotelo JS, López-Fuentes A, Porrón-Sotelo L, Alquicira-Hernández S, Medina-Rivera A, Martínez-Flores I, Alquicira-Hernández K, Martínez-Adame R, Bonavides-Martínez C, Miranda-Ríos J, Huerta AM, Mendoza-Vargas A, Collado-Torres L, Taboada B, Vega-Alvarado L, Olvera M, Olvera L, Grande R, Morett E, Collado-Vides J (2011) RegulonDB version 7.0: transcriptional regulation of Escherichia coli K-12 integrated within genetic sensory response units (Gensor Units). Nucleic Acids Res 39(suppl 1):D98–D105
Gong JX, Zheng HJ, Wu ZJ, Chen T, Zhao XM (2009) Genome shuffling: progress and applications for phenotype improvement. Biotechnol Adv 27(6):996–1005
Gunasekera TS, Csonka LN, Paliy O (2008) Genome-wide transcriptional responses of Escherichia coli K-12 to continuous osmotic and heat stresses. J Bacteriol 190(10):3712–3720
Harman JG (2001) Allosteric regulation of the cAMP receptor protein. Biochim Biophys Acta Prot Struct Mol Enzym 1547(1):1–17
Harry E, Monahan L, Thompson L (2006) Bacterial cell division: the mechanism and its precison. Int Rev Cytol Surv Cell Biol 253:27–94
Hayashi S, Aono R, Hanai T, Mori H, Kobayashi T, Honda H (2003) Analysis of organic solvent tolerance in Escherichia coli using gene expression profiles from DNA microarrays. J Biosci Bioeng 95(4):379–383
Hermann M, Fayolle F, Marchal R, Podvin L, Sebald M, Vandecasteele JP (1985) Isolation and characterization of butanol-resistant mutants of Clostridium acetobutylicum. Appl Environ Microbiol 50(5):1238–1243
Hobert O (2008) Gene regulation by transcription factors and microRNAs. Science 319(5871):1785–1786
Hong SH, Lee J, Wood TK (2010a) Engineering global regulator Hha of Escherichia coli to control biofilm dispersal. Microb Biotechnol 3(6):717–728
Hong SH, Wang X, Wood TK (2010b) Controlling biofilm formation, prophage excision and cell death by rewiring global regulator H-NS of Escherichia coli. Microb Biotechnol 3(3):344–356
Jeong H, Han J (2010) Enhancing the 1-butanol tolerance in Escherichia coli through repetitive proton beam irradiation. J Korean Phys Soc 56(6):2041–2045
Jia KZ, Zhang YP, Li Y (2010) Systematic engineering of microorganisms to improve alcohol tolerance. Eng Life Sci 10(5):422–429
Jiang RR, Bommarius AS (2004) Hydrogen peroxide-producing NADH oxidase (nox-1) from Lactococcus lactis. Tetrahedron-Asymmetry 15(18):2939–2944
Jiang RR, Riebel BR, Bommarius AS (2005) Comparison of alkyl hydroperoxide reductase (AhpR) and water-forming NADH oxidase from Lactococcus lactis ATCC 19435. Adv Synth Catal 347(7–8):1139–1146
Kern A, Tilley E, Hunter IS, Legisa M, Glieder A (2007) Engineering primary metabolic pathways of industrial micro-organisms. J Biotechnol 129(1):6–29
Keseler IM, Collado-Vides J, Santos-Zavaleta A, Peralta-Gil M, Gama-Castro S, Muniz-Rascado L, Bonavides-Martinez C, Paley S, Krummenacker M, Altman T, Kaipa P, Spaulding A, Pacheco J, Latendresse M, Fulcher C, Sarker M, Shearer AG, Mackie A, Paulsen I, Gunsalus RP, Karp PD (2011) EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res 39:D583–D590
Khankal R, Chin J, Ghosh D, Cirino P (2009) Transcriptional effects of CRP* expression in Escherichia coli. J Biol Eng 3(1):13
Klein-Marcuschamer D, Stephanopoulos G (2008) Assessing the potential of mutational strategies to elicit new phenotypes in industrial strains. Proc Natl Acad Sci U S A 105(7):2319–2324
Klein-Marcuschamer D, Santos CNS, Yu HM, Stephanopoulos G (2009) Mutagenesis of the bacterial RNA polymerase alpha subunit for improvement of complex phenotypes. Appl Environ Microbiol 75(9):2705–2711
Knoshaug EP, Zhang M (2009) Butanol tolerance in a selection of microorganisms. Appl Biochem Biotechnol 153(1–2):13–20
Kongpol A, Pongtharangkul T, Kato J, Honda K, Ohtake H, Vangnai AS (2009) Characterization of an organic-solvent-tolerant Brevibacillus agri strain 13 able to stabilize solvent/water emulsion. FEMS Microbiol Lett 297(2):225–233
Lee SK, Chou H, Ham TS, Lee TS, Keasling JD (2008) Metabolic engineering of microorganisms for biofuels production: from bugs to synthetic biology to fuels. Curr Opin Biotechnol 19(6):556–563
Lin YL, Blaschek HP (1983) Butanol production by a butanol-tolerant strain of Clostridium acetobutylicum in extruded corn broth. Appl Environ Microbiol 45(3):966–973
Liu HM, Yan M, Lai CG, Xu L, Ouyang PK (2010) gTME for improved xylose fermentation of Saccharomyces cerevisiae. Appl Biochem Biotechnol 160(2):574–582
Lountos GT, Jiang RR, Wellborn WB, Thaler TL, Bommarius AS, Orville AM (2006) The crystal structure of NAD(P)H oxidase from Lactobacillus sanfranciscensis: Insights into the conversion of O-2 into two water molecules by the flavoenzyme. Biochemistry 45(32):9648–9659
Luque R, Herrero-Davila L, Campelo JM, Clark JH, Hidalgo JM, Luna D, Marinas JM, Romero AA (2008) Biofuels: a technological perspective. Energy Environ Sci 1(5):542–564
Ma HW, Kumar B, Ditges U, Gunzer F, Buer J, Zeng AP (2004) An extended transcriptional regulatory network of Escherichia coli and analysis of its hierarchical structure and network motifs. Nucleic Acids Res 32(22):6643–6649
Morawski B, Quan S, Arnold FH (2001) Functional expression and stabilization of horseradish peroxidase by directed evolution in Saccharomyces cerevisiae. Biotechnol Bioeng 76(2):99–107
Neumann G, Veeranagouda Y, Karegoudar TB, Sahin O, Mausezahl I, Kabelitz N, Kappelmeyer U, Heipieper HJ (2005) Cells of Pseudomonas putida and Enterobacter sp adapt to toxic organic compounds by increasing their size. Extremophiles 9(2):163–168
Nielsen DR, Leonard E, Yoon S-H, Tseng H-C, Yuan C, Prather KLJ (2009) Engineering alternative butanol production platforms in heterologous bacteria. Metab Eng 11(4–5):262–273
Okochi M, Kurimoto M, Shimizu K, Honda H (2007) Increase of organic solvent tolerance by overexpression of manXYZ in Escherichia coli. Appl Microbiol Biotechnol 73(6):1394–1399
Okochi M, Kanie K, Kurimoto M, Yohda M, Honda H (2008) Overexpression of prefoldin from the hyperthermophilic archaeum Pyrococcus horikoshii OT3 endowed Escherichia coli with organic solvent tolerance. Appl Microbiol Biotechnol 79(3):443–449
Park KS, Lee DK, Lee H, Lee Y, Jang YS, Kim YH, Yang HY, Lee SI, Seol W, Kim JS (2003) Phenotypic alteration of eukaryotic cells using randomized libraries of artificial transcription factors. Nat Biotechnol 21(10):1208–1214
Passner JM, Schultz SC, Steitz TA (2000) Modeling the cAMP-induced allosteric transition using the crystal structure of CAP-cAMP at 2.1 angstrom resolution. J Mol Biol 304(5):847–859
Pianetti A, Battistelli M, Citterio B, Parlani C, Falcieri E, Bruscolini F (2009) Morphological changes of Aeromonas hydrophila in response to osmotic stress. Micron 40(4):426–433
Ritz M, Tholozan JL, Federighi M, Pilet MF (2001) Morphological and physiological characterization of Listeria monocytogenes subjected to high hydrostatic pressure. Appl Environ Microbiol 67(5):2240–2247
Rosenberg M, Gutnick D, Rosenberg E (1980) Adherence of bacteria to hydrocarbons—a simple method for measuring cell-surface hydrophobicity. FEMS Microbiol Lett 9(1):29–33
Rui LY, Cao L, Chen W, Reardon KF, Wood TK (2005) Protein engineering of epoxide hydrolase from Agrobacterium radiobacter AD1 for enhanced activity and enantioselective production of (R)-1-phenylethane-1,2-diol. Appl Environ Microbiol 71(7):3995–4003
Rutherford BJ, Dahl RH, Price RE, Szmidt HL, Benke PI, Mukhopadhyay A, Keasling JD (2010) Functional genomic study of exogenous n-butanol stress in Escherichia coli. Appl Environ Microbiol 76(6):1935–1945
Ryu S, Kim J, Adhya S, Garges S (1993) Pivotal role of amino-acid at position-138 in the allosteric hinge reorientation of camp receptor protein. Proc Natl Acad Sci U S A 90(1):75–79
Shen CR, Lan EI, Dekishima Y, Baez A, Cho KM, Liao JC (2011) Driving forces enable high-titer anaerobic 1-butanol synthesis in Escherichia coli. Appl Environ Microbiol 77(9):2905–2915
Shi BH, Xia XH (2003) Morphological changes of Pseudomonas pseudoalcaligenes in response to temperature selection. Curr Microbiol 46(2):120–123
Steen EJ, Chan R, Prasad N, Myers S, Petzold CJ, Redding A, Ouellet M, Keasling JD (2008) Metabolic engineering of Saccharomyces cerevisiae for the production of n-butanol. Microb Cell Factories 7(1):36
Tomas CA, Welker NE, Papoutsakis ET (2003) Overexpression of groESL in Clostridium acetobutylicum results in increased solvent production and tolerance, prolonged metabolism, and changes in the cell’s transcriptional program. Appl Environ Microbiol 69(8):4951–4965
Tuch BB, Li H, Johnson AD (2008) Evolution of eukaryotic transcription circuits. Science 319(5871):1797–1799
Wang L, Wei L, Chen Y, Jiang RR (2010) Specific and reversible immobilization of NADH oxidase on functionalized carbon nanotubes. J Biotechnol 150(1):57–63
Wang L, Xu R, Chen Y, Jiang R (2011) Activity and stability comparison of immobilized NADH oxidase on multi-walled carbon nanotubes, carbon nanospheres, and single-walled carbon nanotubes. J Mol Catal B Enzym 69(3–4):120–126
Wang L, Chen Y, Jiang R (2012) Nanoparticle-supported consecutive reactions catalyzed by alkyl hydroperoxide reductase. J Mol Catal B Enzym 76:9–14
Winkler J, Rehmann M, Kao KC (2010) Novel Escherichia coli hybrids with enhanced butanol tolerance. Biotechnol Lett 32(7):915–920
Won HS, Seo MD, Ko HS, Choi WS, Lee BJ (2008) Interdomain interaction of cyclic AMP receptor protein in the absence of cyclic AMP. J Biochem 143(2):163–167
Yan Y, Liao J (2009) Engineering metabolic systems for production of advanced fuels. J Ind Microbiol Biotechnol 36(4):471–479
Yu SN, Lee JC (2004) Role of residue 138 in the interdomain hinge region in transmitting allosteric signals for DNA binding in Escherichia coli cAMP receptor protein. Biochemistry 43(16):4662–4669
Zhang H, Lountos G, Ching C, Jiang R (2010) Engineering of glycerol dehydrogenase for improved activity towards 1,3-butanediol. Appl Microbiol Biotechnol 88(1):117–124
Zhang H, Chong H, Ching CB, Jiang R (2012) Random mutagenesis of global transcription factor cAMP receptor protein for improved osmotolerance. Biotechnol Bioeng 109(5):1165–1172
Zhao K, Liu MZ, Burgess RR (2005) The global transcriptional response of Escherichia coli to induced sigma(32) protein involves sigma(32) regulon activation followed by inactivation and degradation of sigma(32) in vivo. J Biol Chem 280(18):17758–17768
Acknowledgments
This work is supported by National Research Foundation, Singapore (NRF-CRP5-2009-03).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, H., Chong, H., Ching, C.B. et al. Engineering global transcription factor cyclic AMP receptor protein of Escherichia coli for improved 1-butanol tolerance. Appl Microbiol Biotechnol 94, 1107–1117 (2012). https://doi.org/10.1007/s00253-012-4012-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-012-4012-5