Abstract
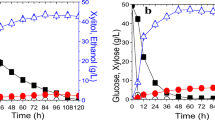
Global transcription machinery engineering (gTME) is an approach for reprogramming gene transcription to elicit cellular phenotypes important for technological applications. In our study, the application of gTME to Saccharomyces cerevisiae was to improve xylose utilization and tolerance, which is a key trait for many biofuel programs. Mutation of the transcription factor spt15 was introduced by error-prone polymerase chain reaction and then screened on media using xylose as the sole carbon source. The selected out strain spt15-25 showed modest growth rates in the media containing 50, 100, and 150 g/L of xylose or glucose. Under the following fermentation condition: 30 °C, rotating speed of 200 r/min, 500-mL Erlenmeyer flask containing 100-mL media, after 109 h, 93.5% of xylose was consumed in 50 g/L xylose medium. Meanwhile, 98.3% glucose can be metabolized in 50-g/L glucose medium. And the carbon source was 50 g/L glucose–xylose (w/w = 1); the utilization ratio of xylose and glucose was 90.8% and 97.3%, respectively. And all the xylitol concentration was below 2.48 g/L.








Similar content being viewed by others
Notes
This refers to the utilization rate of 25 g/L glucose in mixed sugar after 50 h which was 97.3% in the paper.
References
Nigam, J. N. (2001). Development of xylose-fermenting yeast Pichia stipitis for ethanol production through adaptation on hardwood hemicellulose acid prehydrolysate. Journal of Applied Microbiology, 90, 208–215. doi:10.1046/j.1365-2672.2001.01234.x.
Slininger, P. J., Bothast, R. J., Van Cauwenberge, J. E., et al. (1982). Conversion of D-xylose to ethanol by the yeast Pachysolen tannophilus. Biotechnology and Bioengineering, 14, 37–384.
Tang, Y., An, M., Liu, K., et al. (2006). Ethanol production from acid hydrolysate of wood biomass using the flocculating yeast Saccharomyces cerevisiae strain KF-7. Process Biochemistry, 41(4), 909–914. doi:10.1016/j.procbio.2005.09.008.
Goffeau, A., Barrell, B. G., Bussey, B., et al. (1996). Life with 6000 genes. Science, 274(5287), 546, 563–567. doi:10.1126/science.274.5287.546.
Batt, C. A., Carvallo, S., Easson, D. D., et al. (1986). Direct evidence for a xylose metabolic pathway in Saccharomyces cerevisiae. Biotechnology and Bioengineering, 28, 549–553. doi:10.1002/bit.260280411.
Alper, H., Moxley, J., Nevoigt, E., et al. (2006). Engineering yeast transcription machinery for improved ethanol tolerance and production. Science, 314(8), 1565–1568. doi:10.1126/science.1131969.
Heluane, H., Spencer, J. F. T., Spencer, D., et al. (1993). Characterization of hybrids obtained by protoplast fusion, between Pachysolen tannophilus and Saccharomyces cerevisiae. Applied Microbiology and Biotechnology, 40, 98–100. doi:10.1007/BF00170435.
Alper, H., Jin, Y.-S., Moxley, J. F., et al. (2005). Identifying gene targets for the metabolic engineering of lycopene biosynthesis in Escherichia coli. Metabolic Engineering, 7, 155–164. doi:10.1016/j.ymben.2004.12.003.
Alper, H., Miyaoku, K., & Stephanopoulos, G. (2005). Construction of lycopene-overproducing E. coli strains by combining systematic and combinatorial gene knockout targets. Nature Biotechnology, 23, 612–616. doi:10.1038/nbt1083.
Alper, H., & Stephanopoulos, G. (2007). Global transcription machinery engineering: A new approach for improving cellular phenotype. Metabolic Engineering, 9, 258–267. doi:10.1016/j.ymben.2006.12.002.
Alper, H., Moxley, J., Nevoigt, E. G. R., et al. (2006). Supporting online material for engineering yeast transcription machinery for improved ethanol tolerance and production. Science, 314, 1565. doi:10.1126/science.1131969.
Hemsley, A., Arnheim, N., Toney, M. D., et al. (1989). A simple method for site-directed mutagenesis using the polymerase chain reaction. Nucleic Acids Research, 17, 6545–6551. doi:10.1093/nar/17.16.6545.
Robert, H., Schidst, I., Andrew, R., et al. (1995). Studies on the transformation of intact yeast cells by the LiAc/SSDNA/PEG procedure. Yeast (Chichester, England), 11, 355–360. doi:10.1002/yea.320110408.
Hallborn, J., Walfidsson, M., Airaksinen, U., et al. (1991). Xylitol production by recombinant Saccharomyces cerevisiae. Bio Technology, 9, 1090–1095.
Acknowledgements
We acknowledge the National Natural Science Foundation of China (No.U0733001) for financial support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, H., Yan, M., Lai, C. et al. gTME for Improved Xylose Fermentation of Saccharomyces cerevisiae . Appl Biochem Biotechnol 160, 574–582 (2010). https://doi.org/10.1007/s12010-008-8431-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-008-8431-9