Abstract
Chitin derivatives, chitosan and substituted chito-oligosaccharides have a wide spectrum of applications ranging from medicine to cosmetics and dietary supplements. With advancing knowledge about the substrate-binding properties of chitinases, enzyme-based production of these biotechnologically relevant sugars from biological resources is becoming increasingly interesting. Fungi have high numbers of glycoside hydrolase family 18 chitinases with different substrate-binding site architectures. As presented in this review, the large diversity of fungal chitinases is an interesting starting point for protein engineering. In this review, recent data about the architecture of the substrate-binding clefts of fungal chitinases, in connection with their hydrolytic and transglycolytic abilities, and the development of chitinase inhibitors are summarized. Furthermore, the biological functions of chitinases, chitin and chitosan utilization by fungi, and the effects of these aspects on biotechnological applications, including protein overexpression and autolysis during industrial processes, are discussed in this review.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chitinases (E.C. 3.2.1.14) hydrolyze the linear polymer chitin, a polysaccharide of β-(1,4) linked N-acetylglucosamine (GlcNAc; 2-acetamino-2-deoxy-β-d-glucose) units. Glycoside hydrolase (GH) family 18 (www.cazy.org) represents an ancient chitinase family found in all kingdoms of life (Funkhouser and Aronson 2007). Filamentous fungi have many different chitinases belonging to GH family 18 (Seidl 2008). The structural scaffold of the fungal cell is composed of chitin and β-(1,3) glucan (Latgé 2007). Fungal chitinases are therefore not only involved in exogenous chitin decomposition but also in fungal cell wall degradation and remodeling. The variability of fungal GH family 18 proteins makes them ideal candidates for the development of enzymes acting on chitinous carbohydrates used in biotechnology. Besides chitinases, GH family 18 in fungi also contains non-chitinolytic enzymes such as endo-β-N-acetylglucosaminidases which can be used for protein deglycosylation (Stals et al. 2010).
This review gives an overview of the diversity of fungal chitinases and their biochemical properties. Recent advances toward understanding the roles of chitinases in fungal biology, as well as substrate-binding properties and new mechanistic insights in different fungal chitinases, including biotechnological implications of these findings, are discussed in this review.
Chitin and GlcNAc-containing carbohydrates
Chitin—the term comes from a Greek word for tunic, a form of clothing worn in ancient Greece—was first discovered in 1811 by Henri Braconnot as a substance occurring in mushrooms (Braconnot 1811). Nowadays, it is known that chitin is highly abundant in nature. There are two allomorphic forms of chitin, namely, α-chitin and β-chitin, which differ in packing and polarities of adjacent chains in successive sheets (Aam et al. 2010; Chen et al. 2010). Fungal cell walls contain α-chitin. The monosaccharide GlcNAc can also be found in a number of heterogeneous polysaccharides related to chitin. Chitosan is a partially deacetylated form of chitin and, thus, a heteropolymer of GlcNAc and d-glucosamine residues. The name chitosan refers to a continuum of soluble polymeric chitin derivatives that can be described and classified according to their acetylation and polymerization parameters (Aam et al. 2010). Chitosan exhibits a variety of interesting physicochemical and biological properties. In contrast to chitin, chitosan is soluble in dilute aqueous acid solutions. This, in combination with its non-toxicity, biocompatibility and biodegradability, makes chitosan and also chito-oligosaccharides (COSs) suitable for their use in numerous applications in agriculture, cosmetics, water treatment and medicine (Aam et al. 2010; Chen et al. 2010). Chitosan and COSs can be prepared either chemically by acid hydrolysis or enzymatically by hydrolysis with glycosyl hydrolases like chitinases or chitosanases. The resulting COS-mixture depends on the chitosan/chitin starting material and the specificity of the enzyme(s) used. Other natural, heterogeneous polysaccharides that contain GlcNAc are murein, a component of bacterial cell walls; hyaluronic acid, found in connective, epithelial and neural tissues; and chondroitin, which is present in connective tissues, blood vessels, bone and cartilage (Chen et al. 2010).
Enzymes acting on chitin and chitosan
Chitin can be enzymatically cleaved by two main enzyme classes: chitinases and N-acetylglucosaminidases. Furthermore, chitin can be deacetylated by chitin deacetylases (EC 3.5.1.31) found in carbohydrate esterase family 4 (CE 4) in the Carbohydrate Active Enzymes database (CAZy) classification. Until now, chitinases (EC 3.2.1.14) were mainly described from GH families 18 and 19 (Bhattacharya et al. 2007; Fukamizo 2000; Kasprzewska 2003; Schrempf 2001). Fungal chitinases were, so far, exclusively found in GH family 18 (see the following for details). Chitinases are the only enzymes that can efficiently degrade polymeric chitin by cleaving it into COSs with a minimum chain length of n = 2. These products can be further converted to GlcNAc monomers by N-acetylglucosaminidases (EC 3.2.1.52), which in fungi have, so far, only been described from GH family 20. In analogy to chitin, chitosan can be degraded by chitosanases (EC 3.2.1.132) and glucosaminidases (3.2.1.165). Fungal chitosanases belong to GH family 75 (Li et al. 2008; Rodriguez-Martin et al. 2010; Zhang et al. 2001; Cheng et al. 2006), and a glucosaminidase from GH family 2 has been reported from Trichoderma reesei (Ike et al. 2006). Chitinases can also show enzymatic activity on chitosan, but this activity is strongly dependent on the degree of acetylation. This is due to a requirement of chitinases for GlcNAc in the −1 subsite for catalytic cleavage to occur (see ‘Mechanistic properties of fungal chitinases’ for details).
Numbers and modular architecture of fungal chitinases
In filamentous fungi, chitin is located in the inner layers of the cell wall, close to the plasma membrane (Ruiz-Herrera 1991). It is therefore not easily accessible from the outer side of the cell wall as it is masked by layers of other carbohydrates and proteins. Nevertheless, all filamentous fungi have many different chitinases. The chitin content of fungi varies strongly among different fungal species and is dependent on the growth form. Yeast cell walls have a rather low chitin content (0.5–5%). In yeasts, chitin is mainly found in septa, constriction rings and budding scars (Chaffin et al. 1998; Bulik et al. 2003). Cell walls of filamentous fungi consist of up to 20% or more of chitin, which can be found throughout the whole cell wall of hyphae (Ruiz-Herrera 1991). In general, spore walls contain lesser amounts of chitin than hyphae. Interestingly, the chitin content of fungi is reflected in their numbers of chitinases. Yeasts and fungi with yeast-like growth forms have low numbers of chitinases—Saccharomyces cerevisiae has two, Candida albicans has four, and the dimorphic basidiomycete Ustilago maydis has two—but filamentous fungi have, in general, between 10 and 20 different GH family 18 proteins (Seidl 2008). Mycoparasitic and entomopathogenic fungi have even 30 or more GH family 18 proteins (Gao et al. 2011; Kubicek et al. 2011). It should be noted that for most of these proteins, biochemical evidence verifying that they are active chitinases is still missing. However, based on aa-similarities and the presence of conserved aa-residues that are essential for catalytic cleavage, it can be assumed that most of these GH family 18 proteins are indeed chitinases (Gruber et al. 2011b; Seidl et al. 2005).
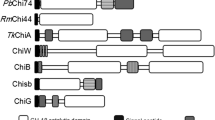
Fungal chitinases can be divided into three different subgroups, namely, A, B and C, based on the amino acid sequences of their GH 18 modules. These subgroups differ in the architectures of their substrate-binding cleft and, thus, their enzymatic activities (exo vs. endo) and also contain different carbohydrate-binding modules (CBM 18 and 50; see also www.cazy.org; Gruber and Seidl-Seiboth 2011; Seidl 2008). The properties of chitinase subgroups A–C are summarized in Table 1. The presence of CBMs in enzymes enables them to bind more tightly to insoluble substrates (Eijsink et al. 2008). Furthermore, processivity of enzymes can be enhanced by CBMs.
Mechanistic properties of fungal chitinases
As shown in Table 1, fungal sg A and C chitinases belong to class V (= fungal/bacterial) chitinases, and sg B belongs to class III (= fungal/plant) chitinases. Class III chitinases, e.g. the plant chitinase hevamine, have a shallow and open active site architecture indicative for endochitinases (Terwisscha van Scheltinga et al. 1996). In contrast, class V chitinases, e.g. the bacterial chitinases ChiA and ChiB from Serratia marcescens and the fungal chitinase (sg A) CiX1 from Coccidioides immitis, have a deep and tunnel-shaped active site and are exochitinases (Bortone et al. 2002; van Aalten et al. 2000). In the substrate-binding cleft, several sugars from a chain of the chitin polymer or a COS can be accommodated. In general, the substrate-binding site of GH family 18 chitinases is relatively long and accommodates at least five sugar units. The sugar-binding subsites are denominated as −3, -2, -1, +1, +2 and +3, and cleavage occurs between the −1 and +1 sugar (Fig. 1). All GH family 18 proteins have a (α/β)8 barrel (TIM barrel) fold. The substrate-binding amino acids are located in loops extending from the (α/β)8 barrel fold, and in some chitinases, additional interactions are caused by an extra domain existing as a barrel insertion (Lienemann et al. 2009). GH family 18 chitinases are retaining enzymes, which means that the β-anomeric configuration found in the substrate is retained in the product. GH family 18 chitinases have a substrate-assisted catalytic mechanism (Bortone et al. 2002; van Aalten et al. 2001). In this reaction mechanism, the acid protonating the glycosidic bond is a conserved glutamate, and the nucleophile is the oxygen of the N-acetyl group on the −1 sugar, forming an oxazolinium ion intermediate. Due to this unusual enzymatic mechanism, a GlcNAc residue in the −1 subsite is mandatory for catalytic cleavage to occur. The (sugar-) binding requirements at subsites more distal from the cleavage point are more flexible, and therefore, different sugar compositions are tolerated. This type of flexibility is a good starting point for directed evolution and protein-engineering approaches to improve substrate specificity toward biotechnologically relevant sugars, e.g. animal-like β-1,4-galactosylated and α-1,3-fucosylated chito-oligosaccharides (Boer et al. 2004).
Schematic representation of the substrate-binding clefts of GH family 18 chitinases. a Chitinases have multiple sugar-binding sites in their long substrate-binding clefts. Cleavage occurs between the +1 and −1 sugar. In these two subsites, the substrate has contact with two amino acids (D and E) that are part of the diagnostic catalytic DXXDXDE motif, which is essential for catalytic cleave by GH family 18 chitinases. b Sg A (class V) and C chitinases have narrow and tunnel-shaped substrate-binding clefts, and sg B (class III) chitinases have wide and open substrate-binding clefts
Toward the utilization of modified COSs structural modeling and enzymatic characterization of the two chitinases Chit42 and Chit33 from Trichoderma harzianum, members of sg A and B, respectively, were carried out (Boer et al. 2007; Lienemann et al. 2009). This also enabled a direct comparison of representative members of these two subgroups. Chit42 and Chit33 were characterized using a panel of different soluble chitinous substrates and inhibitors. Chit42 has at least seven subsites (−5 to +2), and Chit33 has at least six (−4 to +2). Both chitinases cleave preferentially between the second and third sugar from the reducing end of the substrate, but they have different kinetic behaviors. While Chit42 can degrade chitohexaose (GlcNAc)6 in a processive manner into three (GlcNAc)2 units, Chit33 produces (GlcNAc)2 and (GlcNAc)4, which are not further degraded. This is in agreement with a preferential exo/endo-mode of related chitinases and is further supported by the modeled structures. Chit42 has a deep substrate-binding groove, as has also been found for Coccidioides immitis CiX1 and bacterial GH family 18 (class V) exochitinases. Chit33, on the other hand, has a more shallow and open substrate-binding site, similar to related plant (class III) endochitinases. However, in this context, it should be noted that the strict differentiation between exo- and endochitinases can be difficult because chitinases that cleave preferentially in an exo-mode can also have endo-activity and vice versa. Detailed biochemical analyses of more fungal chitinases will be necessary to elucidate the spectrum of cleavage patterns in different chitinases.
Chit42 variants with altered substrate specificities were created based on the obtained enzyme–substrate interaction data from modeling experiments (Lienemann et al. 2009). Alteration of either charged or hydrophobic interactions between the enzyme and the substrate, particularly in the −3 subsite, changed and/or broadened the substrate preferences of Chit42. This shows that the long binding site groove of Chit42 offers possibilities for modification of the enzyme toward, for example, pharmaceutically relevant oligosaccharides, e.g. COSs with defined acetylation patterns or substitutions at defined positions.
Another sg B chitinase that was already enzymatically characterized in more detail is Trichoderma atroviride Ech30 (Chi18-13; Hoell et al. 2005). Ech30 is a small chitinase (30 kDa) but has as many as seven subsites for sugar binding. While the well-characterized plant chitinase hevamine has six subsites, running from −4 to +2, Ech30 does not seem to have a −4 subsite, but a sugar-binding site at +3 and maybe even +4. Structural modeling showed that this is due to subtle differences in the substrate-binding cleft, resulting from small insertions and deletions in loops on the non-catalytic side of the TIM barrel. These data underline the large variability and yet unexploited potential of different chitinases for the development of biotechnological applications.
So far, no structural or biochemical data are available about fungal sg C chitinases. Based on structural modeling and protein alignments, sg C chitinases are distantly related to sg A chitinases and also have rather narrow and deep substrate-binding sites (Gruber et al. 2011b). Due to their multiple CBMs—CBM 18 (chitin binding) and CBM 50 (lysin motif; LysM)—the substrate-binding properties of sg C chitinases might be different from sg A chitinases, which have no CBMs. The LysM is an ancient protein domain originally identified in bacterial autolysin (Joris et al. 1992). The presence of CBM 50 (LysM) domains in chitinases is especially interesting because bacterial proteins with LysM domains have been reported to be involved in specific recognition events between nitrogen fixing bacteria and their plant hosts (Knogge and Scheel 2006). This recognition is mediated by nodulation (Nod) factors, which are lipo-COSs with several functional group substitutions, including sulfation and fucosylation. The aa-sequences of fungal LysM domains are highly variable, and they could therefore be important for substrate-binding specificity and, thus, targeting of the chitinase to its correct place for hydrolysis.
Transglycosylation
GH family 18 chitinases are also able to catalyze transglycosylation reactions. Transglycosylation products were already reported for T. harzianum Chit42 and Chit33, as well as for Aspergillus fumigatus ChiB1 (Andres et al. 2011; Boer et al. 2004; Lü et al. 2009). In ChiB1, mutation of aa-residues involved in substrate-binding altered hydrolytic and transglycosyl activities of this enzyme (Lü et al. 2009). In the case of Chit42 and Chit33, transglycosylation was also observed with synthetically modified COSs, underlining the potential applications of these enzymes. Biotechnologically, these types of enzymatic transglycosylation reactions are of interest because the chemical synthesis of complex oligosaccharides with substitutions at defined positions is usually very tedious. Following the principle of the ‘glycosynthase technology’ in which glycosidases are engineered to catalyze transglycosylation reactions (Shaikh and Withers 2008), conserved catalytic residues of Chit42 were mutated and screened for transglycosylation behavior (Andres et al. 2011). Enzymes in which the hydrolytic activity was abolished, but that catalyzed coupling reactions between substituted and non-substituted COSs, were successfully generated by mutation of a stabilizing aspartate. The use of enzymes for these purposes would significantly increase the selectivity of the synthesis reaction due to the different carbohydrate–protein interactions at various sugar-binding subsites.
Chitinase inhibitors
Chitinase inhibitors have a chemotherapeutic potential against fungi due to the proposed roles of chitinases in chitin remodeling in the fungal cell wall (Rao et al. 2005). Several different classes of chitinase inhibitors have been reported. The most potent natural inhibitors are the pseudotrisaccharide allosamidin and the cyclic pentapeptides argifin and argadin (Fig. 2 a–c). Allosamidin is a transition state analog and a strong inhibitor of family GH 18 chitinases. Unfortunately, total synthesis of allosamidin is difficult and expensive, making it not a suitable candidate for further optimization as a chemotherapeutical agent (Andersen et al. 2005).
The two cyclic pentapeptides argifin and argadin were the first non-sugar molecules that were reported to inhibit chitinases in the submicromolar range (Andersen et al. 2005). Both of these inhibitors were isolated from fungi. Argifin was extracted from mycelia of Gliocladium sp., and argadin was extracted from mycelia of Clonostachys sp. (Arai et al. 2000; Omura et al. 2000). Fungal species identification was based on morphological characteristics. Several fungal species from both of these genera are well-known entomopathogens, mycoparasites and/or nematophagous fungi. However, the taxonomical classification of these fungi based on morphological parameters can be difficult. The use of DNA-based phylogenetic markers in the past decades therefore led to the reclassification and renaming of numerous fungal species. For example, Trichoderma virens and Clonostachys rosea were formerly known as Gliocladium species (Rehner and Samuels 1994; Schroers et al. 1999). Thus, a taxonomic reassessment with DNA-based phylogenetic markers of the two species from which the chitinase inhibitors argifin and argadin were derived would be interesting. Furthermore, these phylogenetic relations show that fungi with an aggressive lifestyle, such as mycoparasites or entomopathogens, might be a yet unexploited resource for novel chitinase inhibitors.
Argifin and argadin occupy the −1, +1 and +2 subsites in the active groove of chitinases (Houston et al. 2002). These inhibitors act on human, bacterial and fungal chitinases, but subtle changes in the binding site dramatically affect affinity and selectivity. The crystal structure of A. fumigatus ChiB1 (sg A) was solved in complex with argifin and argadin (Rao et al. 2005). Although argifin and argadin share a similar structure, the dissociation constant of argifin to A. fumigatus ChiB1 is in the nanomolar range, but that of argadin is only in the micromolar range.
A low molecular weight compound composed of two linked caffeine moieties (Fig. 2d), derived from a screening-based approach, was shown to act as a competitive inhibitor on ChiB1 in the low micromolar (K i = 2.8 ± 0.2 μm) range (Schüttelkopf et al. 2006). Small chitinase inhibitors of this type provide attractive, synthetically accessible scaffolds for further optimization. Toward the development of inhibitors for sg B chitinases with open substrate-binding clefts, a small, highly efficient, argifin-derived, nine-atom fragment was shown to be a micromolar inhibitor ChiA1 (Rush et al. 2010). These recent advances show that the synthesis of novel chitinase inhibitors has attracted considerable interest due to their wide potential applications in medicine.
Biological functions of chitinases in filamentous fungi
In contrast to many other carbohydrate-active enzymes that are solely involved in substrate degradation for nutritional purposes, chitinases have multiple functions in fungal biology (Seidl 2008). For a rational design-based development of chitinases for biotechnological applications, an integrated understanding of the different roles of chitinases and, thus, their natural substrates will be necessary.
Subgroup A seems to contain chitinases involved in processes during fungal growth and autolysis. Autolysis is the natural process of self-digestion of aged hyphal cultures. A better understanding of the mechanisms and factors causing fungal autolysis has the potential for considerable improvements in various fungal applications. In bioprocessing industries, autolysis can have a pronounced effect on product yields (White et al. 2002). In some biotechnological processes, autolysis needs to be prevented to continue antibiotic production or prevent degradation of heterologous proteins. In other cases, autolysis can be advantageous because it enables the release of intracellular products or facilitates downstream processing. Another area where autolysis might be of relevance is the treatment of fungal infections, which aims at the death of the fungus.
Most fungal chitinases that were characterized so far phylogenetically belong to the same clade of sg A and are involved in autolysis in various fungi (Aspergillus nidulans ChiB, A. fumigatus ChiB1, Penicillium chrysogenum PcChiB1, T. atroviride Ech42, T. harzianum Chit42 and T. virens Cht42/Tv-Ech1). However, the contribution of these different chitinases to autolysis interestingly is strongly variable among the different fungi (Shin et al. 2009; Jaques et al. 2003; Kamerewerd et al 2011). T. atroviride Ech42 is involved in autolysis as well as in mycoparasitism. As has been discussed in a recent review (Gruber and Seidl-Seiboth 2011), these data suggest that the same chitinases can participate in self- and non-self cell wall degradation. It was suggested that the accessibility of chitin within the fungal cell wall could be a major determinant in these processes. In healthy hyphae, accessibility may be limited by protection conferred by cell wall proteins. This is supported by the finding that germination of T. atroviride spores is not inhibited by an enzyme mixture containing chitinolytic enzymes from the autolytic phase of T. atroviride cultures. In contrast, germination of other fungi (T. reesei, Neurospora crassa and Aspergillus niger) is strongly impaired by these enzymes (Fig. 3). For other genes from sg A, basal expression levels under all tested growth conditions were detected, indicating an involvement of the proteins in hyphal development and colony formation.
Effect of a chitinolytic enzyme mix from the autolytic phase of T. atroviride cultures on germination of different fungi. a T. atroviride. b T. reesei. c Neurospora crassa. d A. niger. Spores were incubated in potato dextrose broth and at 28° for 15 h (Ta, Tr, An) or 8 h (Nc). In 20-mM Tris–HCl (pH 8.0) buffer or buffer alone (control), 33% of the medium were replaced with T. atroviride enzymes. Scale bars = 20 μm
Subgroup B chitinases appear mainly to be involved in nutritional functions, including more aggressive functions in, for example, mycoparasitic and entomopathogenic fungi. Most tested sg B chitinase genes from other fungal species are regulated by nutritional stimuli. In general, they are inducible by chitin or more host-specific carbon sources in mycoparasitic and entomopathogenic fungi. Most of them are also upregulated during starvation (e.g. T. atroviride chit33 and ech30 and M. anisopliae chi2) and are repressed by glucose or other easily metabolizable carbon sources (de las Mercedes Dana et al. 2001; Klemsdal et al. 2006; Seidl et al. 2005; Yamazaki et al. 2008; Baratto et al. 2006; da Silva et al. 2005). Knock-out strains of chi2 showed decreased virulence to insects (i.e. the cotton stainer bug Dysdercus peruvianus) (Boldo et al. 2009).
Yeast chitinases—Saccharomyces cerevisiae Cts1p and Candida albicans CHT2 and CHT3—that are related to sg B chitinases of filamentous fungi are required for cell separation (Dünkler et al. 2005; Kuranda and Robbins 1991). The glycosylphosphatidylinositol (GPI)-anchored chitinase ChiA of A. nidulans localizes at the germ tubes of conidia, hyphal branching sites and hyphal tips (Yamazaki et al. 2008). This led to the functional analysis of all five A. fumigatus sg B chitinases by generating a quintuple knockout strain (Alcazar-Fuoli et al. 2011). Only a limited reduction in the total chitinolytic activity was detected for the different chitinase mutants, including the quintuple mutant, and no growth or germination defects were observed. These results demonstrate that sg B chitinases—including a GPI-anchor containing orthologue of A. nidulans ChiA—do not have an essential role in morphogenesis in A. fumigatus.
Subgroup C chitinases were, so far, mainly investigated in the two mycoparasites T. atroviride and T. virens. Completely different transcript patterns were obtained in these two fungi, even for genes that are > 90% identical at the DNA level. In T. atroviride, mycoparasitism-related growth conditions induced all sg C chitinase genes in a concerted manner (Gruber et al. 2011b). Only T. atroviride tac6, which probably encodes a chitolectin, was expressed during hyphal network formation. Starvation did not induce sg C chitinases in T. atroviride. In contrast, in T. virens, complex sg C chitinase gene expression patterns were obtained during all growth conditions. These patterns were not only dependent on nutritional stimuli but also on the hyphal zone that was harvested and on the mode of cultivation (Gruber et al. 2011a). These data showed that developmental and morphogenetic signals derived from inter- and intrahyphal interactions regulate sg C chitinase expression in T. virens. Different growth morphologies were observed in T. atroviride and T. virens. It therefore was suggested that hyphal interaction and network formation could be relevant for sg C chitinase gene regulation. Based on the so far available data, it can be concluded that sg C chitinases are involved in several aspects of self- and non-self chitin degradation.
Chitin and chitosan utilization in fungi
Chitin has been frequently used as carbon source for fungi that have many chitinases, e.g. mycoparasites. However, although mycoparasitic Trichoderma spp. have many chitinolytic enzymes, chitin is not a good substrate for these fungi (Gruber et al. 2011a). In contrast to the entomopathogen Beauveria bassiana or the nematophagous fungus Pochonia chlamydosporia, growth of Trichoderma spp. on media containing chitosan is strongly impaired (Palma-Guerrero et al. 2008). Chitosan causes fungal death by permeabilizing the plasma membrane of fungi (Palma-Guerrero et al. 2010). Different susceptibilities toward chitosan between sensitive and non-sensitive fungi have been attributed to differences in membrane fluidity. Pochonia chlamydosporia showed large clearing zones on agar plates amended with chitosan, indicating high amounts of secreted, chitosanolytic enzmes (Palma-Guerrero et al. 2008). This was not observed for Trichoderma spp. It is possible that fungal parasites of invertebrates also show a better capability to metabolize pure chitin than mycoparasitic Trichoderma spp., but this has not been directly compared yet. However, it is interesting to note that the growth data of T. atroviride on chitin and chitosan are somewhat in contrast to the numbers of chitinolytic and chitosanolytic enzymes that are encoded in the genomes of mycoparasitic Trichoderma spp. These results indicate that T. atroviride is not adapted to the utilization of these carbon sources, which it probably does not encounter frequently in its natural habitat in their pure form, for nutritional purposes. The poor growth of T. atroviride on chitin shows that the large number of different chitinases probably rather reflects a chitinolytic system that can degrade a wide range of different chitinous substrates than the good degradation capacity of bulk chitin. A versatile chitin degradation machinery is potentially also less susceptible to chitinase inhibitors, proteases and other adverse components that the fungus might encounter in nature. However, the growth data of T. atroviride on chitin indicate that a large chitinolytic system does not necessarily reflect a good chitin degradation potential. Consequently, for large-scale production of fungal chitinases with selected, biotechnologically relevant properties, overexpression in heterologous systems will probably be necessary.
Overexpression of fungal chitinases for biotechnological applications
There are two types of overexpression of fungal chitinases: (a) overexpression in biological systems for making use of the chitinase activity in vivo and (b) overexpression of chitinases for enzyme-based utilization of the chitinase in vitro.
Constitutive overexpression of antifungal genes from microorganisms involved in plant defense mechanisms in agriculturally important plants represents a promising strategy for conferring genetic resistance against a broad range of plant pathogenic fungi. Expression of chitinase encoding genes in plants has been shown to improve their defense response against various fungal pathogens. In particular, chitinases from mycoparasitic Trichoderma spp. were already overexpressed in several agriculturally important plants, e.g. lemon, cotton, apple and carrot (de las Mercedes Dana et al. 2006; Kumar et al. 2009 and references therein). Transgenic plants, in addition to having a shield provided by the transgenic chitinase, are, in general, at a heightened level of defense as manifested by higher activities of various defense-related endogenous genes and enzymes. Furthermore, these plants launch the defense-related response more rapidly compared to the non-transgenic plants. Besides an increased resistance against fungal pathogens, transgenic tobacco plants (Nicotiana tabacum) overexpressing chitinases were also shown to be more resistant against other types of abiotic and biotic stresses such as bacterial pathogens, salinity and heavy metals (de las Mercedes Dana et al. 2006). This is probably mediated by defense-related mechanisms which are being triggered by the presence of the chitinase in the apoplast. Such broad-range protective effects interestingly came off with no obvious detrimental effect on the growth of tobacco plants. The phenotype of the chitinase-overexpressing plants was morphologically indistinguishable from that of control lines with regard to biomass production, fertility and seed viability. Although these results are promising, from a biotechnologically applied point of view, it remains to be seen whether products from genetically engineered plants would become accepted in the market. Currently, legal restrictions and customer preferences do not (strongly) support the use of genetically modified plants.
In aggressive fungi, e.g. mycoparasites and parasites of invertebrates, chitinases are involved in the attack of other fungi and insects, respectively. Overexpression of single chitinases in mycoparasitic and entomopathogenic fungi and/or addition of a chitin-binding domain to these chitinases was shown to improve the mycoparasitic potential and insect virulence, respectively, of these fungi (Boldo et al. 2009; Fan et al. 2007a; Limon et al. 1999). However, in analogy to the issue with plants mentioned previously, although these results aid in advancing our understanding about the roles of chitinases in these processes, it is questionable whether genetically engineered fungi would be suitable biological control agents in agricultural applications.
With respect to large-scale production of fungal chitinases for bioprocessing industries, such as the synthesis of COSs, and high-throughput screening approaches for chitinase mutants, the commonly used heterologous expression system Escherichia coli was already evaluated (Boer et al. 2007; Lienemann et al. 2009). For Chit42 and Chit33, expression in different compartments in E. coli was tested, and periplasmic expression and cell surface display for enzyme screening were investigated. However, this bacterial system proved to be somewhat challenging for the large-scale expression of fungal chitinases due to the presence of disulfide bridges (e.g. Chit33 has two disulfide bridges) or posttranslational modifications such as glycosylation in these proteins. Incorrect folding of enzymes can have an effect on enzyme activity and temperature stability.
T. atroviride Ech42, the orthologue of Chit42 from T. harzianum, was also overexpressed in Pichia pastoris using a constitutive expression system (Perez-Martinez et al. 2007). Enzyme kinetic parameters for the recombinant protein were reported to be identical to those of the enzyme isolated from T. atroviride. Overexpression of B. bassiana Bbchit1—a chitinase that was acquired by horizontal gene transfer from bacteria in the ancestor of the order Hypocreales (Gruber and Seidl-Seiboth 2011)—in Pichia pastoris was shown to improve protein yields and specific activity in comparison to E. coli (Fan et al. 2007b). Eukaryotic systems such as Pichia pastoris or even filamentous fungi, e.g. A. niger or T. reesei, in combination with protein secretion of the overexpressed protein, might therefore be preferable for fungal chitinases due to improved protein folding and glycosylation.
Chitin has been called the most underexploited biomass resource available on Earth (Tharanathan and Kittur 2003). Chitin-based products have a wide range of applications, and several good reviews are available on this topic (Aam et al. 2010; Chen et al. 2010; Tharanathan and Kittur 2003; Ravi Kumar 2000). Applications of GlcNAc include treatment of joint damage, treatment of inflammatory bowel disease, production of cosmetics, sialic acid production, waste water treatment, drug delivery vehicles, etc. Applications of COSs include drugs against asthma, antibacterial agents, ingredients in wound dressings, anticancer therapy, nerve regeneration and vectors in gene therapy. Several other potential effects of COSs have been described, including immune modulatory effects, antifungal activities and lowering effect on serum glucose levels in diabetics. For chemical hydrolysis and deacetylation of chitin, large amounts of HCl, NaOH and water are needed (Ravi Kumar 2000). The development of efficient enzyme-based methods for chitin processing might be more ecologically compatible and reduce requirements for energy and resources of this process. Furthermore, with increasing knowledge about the strong effects that variations in acetylation can have, the demand for COSs with defined parameters increases steadily. An enzymatic synthesis of COSs will facilitate the production of homogenous batches of COSs with defined properties in comparison to chemical treatment and synthesis, which might be too harsh and less specific. As discussed in this review, fungal chitinases constitute a rich arsenal of different proteins, suitable for the development of these applications. A concerted understanding of various aspects of fungal chitinases, ranging from gene regulation and protein functions to biochemical features and substrate-binding properties of different chitinases, will be necessary for advancing chitinase research toward biotechnological applications.
References
Aam BB, Heggset EB, Norberg AL, Sorlie M, Varum KM, Eijsink VG (2010) Production of chitooligosaccharides and their potential applications in medicine. Mar Drugs 8(5):1482–1517. doi:10.3390/md8051482
Alcazar-Fuoli L, Clavaud C, Lamarre C, Aimanianda V, Seidl-Seiboth V, Mellado E, Latge JP (2011) Functional analysis of the fungal/plant class chitinase family in Aspergillus fumigatus. Fungal Genet Biol 48(4):418–429. doi:j.pep/j.fgb.2010.12.007
Andersen OA, Dixon MJ, Eggleston IM, van Aalten DM (2005) Natural product family 18 chitinase inhibitors. Nat Prod Rep 22(5):563–579. doi:10.1039/b416660b
Andres E, Boer H, Koivula A, Samain E, Driguez H, Armand S, Cottaz S (2011) Engineering chitinases for the synthesis of chitin oligosaccharides: catalytic amino acid mutations convert the GH-family 18 glycoside hydrolases into transglycosylases. J Mol Cat B. in press. doi:j.pep/j.molcatb.2011.09.003
Arai N, Shiomi K, Yamaguchi Y, Masuma R, Iwai Y, Turberg A, Kolbl H, Omura S (2000) Argadin, a new chitinase inhibitor, produced by Clonostachys sp. FO-7314. Chem Pharm Bull (Tokyo) 48(10):1442–1446. doi:10.1002/chin.200111186
Baratto CM, Dutra V, Boldo JT, Leiria LB, Vainstein MH, Schrank A (2006) Isolation, characterization, and transcriptional analysis of the chitinase chi2 gene (DQ011663) from the biocontrol fungus Metarhizium anisopliae var. anisopliae. Curr Microbiol 53(3):217–221. doi:10.1007/s00284-006-0078-6
Bhattacharya D, Nagpure A, Gupta RK (2007) Bacterial chitinases: properties and potential. Crit Rev Biotechnol 27(1):21–28. doi:10.1080/07388550601168223
Boer H, Munck N, Natunen J, Wohlfahrt G, Soderlund H, Renkonen O, Koivula A (2004) Differential recognition of animal type β-4-galactosylated and α-3-fucosylated chito-oligosaccharides by two family 18 chitinases from Trichoderma harzianum. Glycobiology 14(12):1303–1313. doi:10.1093/glycob/cwh121
Boer H, Simolin H, Cottaz S, Soderlund H, Koivula A (2007) Heterologous expression and site-directed mutagenesis studies of two Trichoderma harzianum chitinases, Chit33 and Chit42, in Escherichia coli. Protein Expr Purif 51(2):216–226. doi:j.pep/j.pep.2006.07.020
Boldo JT, Junges A, do Amaral KB, Staats CC, Vainstein MH, Schrank A (2009) Endochitinase CHI2 of the biocontrol fungus Metarhizium anisopliae affects its virulence toward the cotton stainer bug Dysdercus peruvianus. Curr Genet 55(5):551–560. doi:10.1007/s00294-009-0267-5
Bortone K, Monzingo AF, Ernst S, Robertus JD (2002) The structure of an allosamidin complex with the Coccidioides immitis chitinase defines a role for a second acid residue in substrate-assisted mechanism. J Mol Biol 320(2):293–302. doi:j.pep/S0022-2836(02)00444-8
Braconnot H (1811) Sur la nature des champignons. Ann Chim Phys 79:265–304
Bulik DA, Olczak M, Lucero HA, Osmond BC, Robbins PW, Specht CA (2003) Chitin synthesis in Saccharomyces cerevisiae in response to supplementation of growth medium with glucosamine and cell wall stress. Eukaryot Cell 2(5):886–900. doi:10.1128/EC.2.5.886-900.2003
Chaffin WL, Lopez-Ribot JL, Casanova M, Gozalbo D, Martinez JP (1998) Cell wall and secreted proteins of Candida albicans: identification, function, and expression. Microbiol Mol Biol Rev 62(1):130–180
Chen JK, Shen CR, Liu CL (2010) N-acetylglucosamine: production and applications. Mar Drugs 8(9):2493–2516. doi:10.3390/md8092493
Cheng CY, Chang CH, Wu YJ, Li YK (2006) Exploration of glycosyl hydrolase family 75, a chitosanase from Aspergillus fumigatus. J Biol Chem 281(6):3137–3144. doi:10.1074/jbc.M512506200
da Silva MV, Santi L, Staats CC, da Costa AM, Colodel EM, Driemeier D, Vainstein MH, Schrank A (2005) Cuticle-induced endo/exoacting chitinase CHIT30 from Metarhizium anisopliae is encoded by an ortholog of the chi3 gene. Res Microbiol 156(3):382–392. doi:j.pep/j.resmic.2004.10.013
de las Mercedes Dana M, Limon MC, Mejias R, Mach RL, Benitez T, Pintor-Toro JA, Kubicek CP (2001) Regulation of chitinase 33 (chit33) gene expression in Trichoderma harzianum. Curr Genet 38(6):335–342. doi:10.1007/s002940000169
de las Mercedes Dana M, Pintor-Toro JA, Cubero B (2006) Transgenic tobacco plants overexpressing chitinases of fungal origin show enhanced resistance to biotic and abiotic stress agents. Plant Physiol 142:722–730. doi:10.1104/pp.106.086140
Dünkler A, Walther A, Specht CA, Wendland J (2005) Candida albicans CHT3 encodes the functional homolog of the Cts1 chitinase of Saccharomyces cerevisiae. Fungal Genet Biol 42(11):935–947. doi:j.pep/j.fgb.2005.08.001
Eijsink VG, Vaaje-Kolstad G, Varum KM, Horn SJ (2008) Towards new enzymes for biofuels: lessons from chitinase research. Trends Biotechnol 26(5):228–235. doi:j.pep/j.tibtech.2008.02.004
Fan Y, Fang W, Guo S, Pei X, Zhang Y, Xiao Y, Li D, Jin K, Bidochka MJ, Pei Y (2007a) Increased insect virulence in Beauveria bassiana strains overexpressing an engineered chitinase. Appl Environ Microbiol 73(1):295–302. doi:10.1128/AEM.01974-06
Fan Y, Zhang Y, Yang X, Pei X, Guo S, Pei Y (2007b) Expression of a Beauveria bassiana chitinase (Bbchit1) in Escherichia coli and Pichia pastoris. Protein Expr Purif 56(1):93–99. doi:j.pep/j.pep.2007.06.012
Fukamizo T (2000) Chitinolytic enzymes: catalysis, substrate binding, and their application. Curr Protein Pept Sci 1(1):105–124
Funkhouser JD, Aronson NN Jr (2007) Chitinase family GH18: evolutionary insights from the genomic history of a diverse protein family. BMC Evol Biol 7:96. doi:10.1186/1471-2148-7-96
Gao Q, Jin K, Ying SH, Zhang Y, Xiao G, Shang Y, Duan Z, Hu X, Xie XQ, Zhou G, Peng G, Luo Z, Huang W, Wang B, Fang W, Wang S, Zhong Y, Ma LJ, St Leger RJ, Zhao GP, Pei Y, Feng MG, Xia Y, Wang C (2011) Genome sequencing and comparative transcriptomics of the model entomopathogenic fungi Metarhizium anisopliae and M. acridum. PLoS Genet 7(1):e1001264. doi:10.1371/journal.pgen.1001264
Gruber S, Seidl-Seiboth V (2011) Self vs. non-self: fungal cell wall degradation in Trichoderma. Microbiology. doi:10.1099/mic.0.052613-0
Gruber S, Kubicek CP, Seidl-Seiboth V (2011a) Differential regulation of orthologous chitinase genes in mycoparasitic Trichoderma species. Appl Environ Microbiol 77(20):7217–7226
Gruber S, Vaaje-Kolstad G, Matarese F, Lopez-Mondejar R, Kubicek CP, Seidl-Seiboth V (2011b) Analysis of subgroup C of fungal chitinases containing chitin-binding and LysM modules in the mycoparasite Trichoderma atroviride. Glycobiology 21(1):122–133. doi:10.1093/glycob/cwq142
Hoell IA, Klemsdal SS, Vaaje-Kolstad G, Horn SJ, Eijsink VG (2005) Overexpression and characterization of a novel chitinase from Trichoderma atroviride strain P1. Biochim Biophys Acta 1748(2):180–190. doi:j.pep/j.bbapap.2005.01.002
Houston DR, Shiomi K, Arai N, Omura S, Peter MG, Turberg A, Synstad B, Eijsink VG, van Aalten DM (2002) High-resolution structures of a chitinase complexed with natural product cyclopentapeptide inhibitors: mimicry of carbohydrate substrate. Proc Natl Acad Sci USA 99(14):9127–9132. doi:10.1073/pnas.132060599
Ike M, Isami K, Tanabe Y, Nogawa M, Ogasawara W, Okada H, Morikawa Y (2006) Cloning and heterologous expression of the exo-beta-d-glucosaminidase-encoding gene (gls93) from a filamentous fungus, Trichoderma reesei PC-3-7. Appl Microbiol Biotechnol 72(4):687–695. doi:10.1007/s00253-006-0320-y
Jaques AK, Fukamizo T, Hall D, Barton RC, Escott GM, Parkinson T, Hitchcock CA, Adams DJ (2003) Disruption of the gene encoding the ChiB1 chitinase of Aspergillus fumigatus and characterization of a recombinant gene product. Microbiology 149(Pt 10):2931–2939. doi:10.1099/mic.0.26476-0
Joris B, Englebert S, Chu CP, Kariyama R, Daneo-Moore L, Shockman GD, Ghuysen JM (1992) Modular design of the Enterococcus hirae muramidase-2 and Streptococcus faecalis autolysin. FEMS Microbiol Lett 70(3):257–264. doi:j.pep/0378-1097(92)90707-U
Kamerewerd J, Zadra I, Kurnsteiner H, Kuck U (2011) PcchiBI, encoding a class V chitinase, is affected by PcVelA and PcLaeA and responsible for cell wall integrity in Penicillium chrysogenum. Microbiology. doi:10.1099/mic.0.051896-0
Kasprzewska A (2003) Plant chitinases—regulation and function. Cell Mol Biol Lett 8(3):809–824
Klemsdal SS, Clarke JL, Hoell IA, Eijsink VG, Brurberg MB (2006) Molecular cloning, characterization, and expression studies of a novel chitinase gene (ech30) from the mycoparasite Trichoderma atroviride strain P1. FEMS Microbiol Lett 256(2):282–289. doi:10.1111/j.1574-6968.2006.00132.x
Knogge W, Scheel D (2006) LysM receptors recognize friend and foe. Proc Natl Acad Sci USA 103(29):10829–10830. doi:10.1073/pnas.0604601103
Kubicek CP, Herrera-Estrella A, Seidl-Seiboth V, Martinez DA, Druzhinina IS, Thon M, Zeilinger S, Casas-Flores S, Horwitz BA, Mukherjee PK, Mukherjee M, Kredics L, Alcaraz LD, Aerts A, Antal Z, Atanasova L, Cervantes-Badillo MG, Challacombe J, Chertkov O, McCluskey K, Coulpier F, Deshpande N, von Doehren H, Ebbole DJ, Esquivel-Naranjo EU, Fekete E, Flipphi M, Glaser F, Gomez-Rodriguez EY, Gruber S, Han C, Henrissat B, Hermosa R, Hernandez-Onate M, Karaffa L, Kosti I, Le Crom S, Lindquist E, Lucas S, Lubeck M, Lubeck PS, Margeot A, Metz B, Misra M, Nevalainen H, Omann M, Packer N, Perrone G, Uresti-Rivera EE, Salamov A, Schmoll M, Seiboth B, Shapiro H, Sukno S, Tamayo-Ramos JA, Tisch D, Wiest A, Wilkinson HH, Zhang M, Coutinho PM, Kenerley CM, Monte E, Baker SE, Grigoriev IV (2011) Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol 12(4):R40. doi:10.1186/gb-2011-12-4-r40
Kumar V, Parkhi V, Kenerley CM, Rathore KS (2009) Defense-related gene expression and enzyme activities in transgenic cotton plants expressing an endochitinase gene from Trichoderma virens in response to interaction with Rhizoctonia solani. Planta 230(2):277–291. doi:10.1007/s00425-009-0937-z
Kuranda MJ, Robbins PW (1991) Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J Biol Chem 266(29):19758–19767
Latgé JP (2007) The cell wall: a carbohydrate armour for the fungal cell. Mol Microbiol 66(2):279–290. doi:10.1111/j.1365-2958.2007.05872.x
Li S, Chen L, Wang C, Xia W (2008) Expression, purification and characterization of endo-type chitosanase of Aspergillus sp. CJ22-326 from Escherichia coli. Carbohydr Res 343(17):3001–3004. doi:j.pep/j.carres.2008.08.032
Lienemann M, Boer H, Paananen A, Cottaz S, Koivula A (2009) Toward understanding of carbohydrate binding and substrate specificity of a glycosyl hydrolase 18 family (GH-18) chitinase from Trichoderma harzianum. Glycobiology 19(10):1116–1126. doi:10.1093/glycob/cwp102
Limon MC, Pintor-Toro JA, Benitez T (1999) Increased antifungal activity of Trichoderma harzianum transformants that overexpress a 33-kDa chitinase. Phytopathology 89(3):254–261. doi:10.1094/PHYTO.1999.89.3.254
Lü Y, Yang H, Hu H, Wang Y, Rao Z, Jin C (2009) Mutation of Trp137 to glutamate completely removes transglycosyl activity associated with the Aspergillus fumigatus AfChiB1. Glycoconj J 26(5):525–534. doi:10.1007/s10719-008-9203-z
Omura S, Arai N, Yamaguchi Y, Masuma R, Iwai Y, Namikoshi M, Turberg A, Kolbl H, Shiomi K (2000) Argifin, a new chitinase inhibitor, produced by Gliocladium sp. FTD-0668. I. Taxonomy, fermentation, and biological activities. J Antibiot (Tokyo) 53(6):603–608. doi:10.1002/chin.200112189
Palma-Guerrero J, Jansson HB, Salinas J, Lopez-Llorca LV (2008) Effect of chitosan on hyphal growth and spore germination of plant pathogenic and biocontrol fungi. J Appl Microbiol 104(2):541–553. doi:10.1111/j.1365-2672.2007.03567.x
Palma-Guerrero J, Lopez-Jimenez JA, Perez-Berna AJ, Huang IC, Jansson HB, Salinas J, Villalain J, Read ND, Lopez-Llorca LV (2010) Membrane fluidity determines sensitivity of filamentous fungi to chitosan. Mol Microbiol 75(4):1021–1032. doi:10.1111/j.1365-2958.2009.07039.x
Perez-Martinez AS, De Leon-Rodriguez A, Harris LJ, Herrera-Estrella A, Barba de la Rosa AP (2007) Overexpression, purification and characterization of the Trichoderma atroviride endochitinase, Ech42, in Pichia pastoris. Protein Expr Purif. doi:j.pep/j.pep.2007.05.009
Rao FV, Houston DR, Boot RG, Aerts JM, Hodkinson M, Adams DJ, Shiomi K, Omura S, van Aalten DM (2005) Specificity and affinity of natural product cyclopentapeptide inhibitors against A. fumigatus, human, and bacterial chitinases. Chem Biol 12(1):65–76. doi:j.pep/j.chembiol.2004.10.013
Ravi Kumar MNV (2000) A review of chitin and chitosan applications. React Funct Polym 46:1–27. doi:j.pep/S1381-5148(00)00038-9
Rehner SA, Samuels GJ (1994) Taxonomy and phylogeny of Gliocladium analyzed by large subunit rDNA sequences. Mycol Res 98:625–634
Rodriguez-Martin A, Acosta R, Liddell S, Nunez F, Benito MJ, Asensio MA (2010) Characterization of the novel antifungal chitosanase PgChP and the encoding gene from Penicillium chrysogenum. Appl Microbiol Biotechnol 88(2):519–528. doi:10.1007/s00253-010-2767-0
Ruiz-Herrera J (1991) Fungal cell wall: structure, synthesis and assembly. CRC Press, Boca Raton
Rush CL, Schuttelkopf AW, Hurtado-Guerrero R, Blair DE, Ibrahim AF, Desvergnes S, Eggleston IM, van Aalten DM (2010) Natural product-guided discovery of a fungal chitinase inhibitor. Chem Biol 17(12):1275–1281. doi:j.pep/j.chembiol.2010.07.018
Schrempf H (2001) Recognition and degradation of chitin by streptomycetes. Antonie Van Leeuwenhoek 79(3–4):285–289. doi:10.1023/A:1012058205158
Schroers HJ, Samuels GJ, Seifert KA, Gams W (1999) Classification of the mycoparasite Gliocladium roseum in Clonostachys as C. rosea, its relationship to Bionectria ochroleuca, and notes on other Gliocladium-like fungi. Mycologia 91:365–385
Schüttelkopf AW, Andersen OA, Rao FV, Allwood M, Lloyd C, Eggleston IM, van Aalten DM (2006) Screening-based discovery and structural dissection of a novel family 18 chitinase inhibitor. J Biol Chem 281(37):27278–27285. doi:10.1074/jbc.M604048200
Seidl V (2008) Chitinases of filamentous fungi: a large group of diverse proteins with multiple physiological functions. Fungal Biol Rev 22:36–42. doi:j.pep/j.fbr.2008.03.002
Seidl V, Huemer B, Seiboth B, Kubicek CP (2005) A complete survey of Trichoderma chitinases reveals three distinct subgroups of family 18 chitinases. FEBS J 272(22):5923–5939. doi:10.1111/j.1742-4658.2005.04994.x
Shaikh FA, Withers SG (2008) Teaching old enzymes new tricks: engineering and evolution of glycosidases and glycosyl transferases for improved glycoside synthesis. Biochem Cell Biol 86(2):169–177. doi:10.1139/O07-149
Shin KS, Kwon NJ, Kim YH, Park HS, Kwon GS, Yu JH (2009) Differential roles of the ChiB chitinase in autolysis and cell death of Aspergillus nidulans. Eukaryot Cell. doi:10.1128/EC.00368-08
Stals I, Samyn B, Sergeant K, White T, Hoorelbeke K, Coorevits A, Devreese B, Claeyssens M, Piens K (2010) Identification of a gene coding for a deglycosylating enzyme in Hypocrea jecorina. FEMS Microbiol Lett 303(1):9–17. doi:10.1111/j.1574-6968.2009.01849.x
Terwisscha van Scheltinga AC, Hennig M, Dijkstra BW (1996) The 1.8 Å resolution structure of hevamine, a plant chitinase/lysozyme, and analysis of the conserved sequence and structure motifs of glycosyl hydrolase family 18. J Mol Biol 262(2):243–257. doi:10.1006/jmbi.1996.0510
Tharanathan RN, Kittur FS (2003) Chitin—the undisputed biomolecule of great potential. Crit Rev Food Sci Nutr 43(1):61–87. doi:10.1080/10408690390826455
van Aalten DM, Synstad B, Brurberg MB, Hough E, Riise BW, Eijsink VG, Wierenga RK (2000) Structure of a two-domain chitotriosidase from Serratia marcescens at 1.9-Å resolution. Proc Natl Acad Sci USA 97(11):5842–5847. doi:10.1073/pnas.97.11.5842
van Aalten DMF, Komander D, Synstad B, Gaseidnes S, Peter MG, Eijsink VGH (2001) Structural insights into the catalytic mechanism of a family 18 exo-chitinase. Proc Natl Acad Sci USA 98(16):8979–8984. doi:10.1073/pnas.151103798
White S, McIntyre M, Berry DR, McNeil B (2002) The autolysis of industrial filamentous fungi. Crit Rev Biotechnol 22(1):1–14. doi:10.1080/07388550290789432
Yamazaki H, Tanaka A, Kaneko J, Ohta A, Horiuchi H (2008) Aspergillus nidulans ChiA is a glycosylphosphatidylinositol (GPI)-anchored chitinase specifically localized at polarized growth sites. Fungal Genet Biol 45(6):963–972. doi:j.pep/j.fgb.2008.02.008
Zhang XY, Dai AL, Kuroiwa K, Kodaira R, Nogawa M, Shimosaka M, Okazaki M (2001) Cloning and characterization of a chitosanase gene from the koji mold Aspergillus oryzae strain IAM 2660. Biosci Biotechnol Biochem 65(4):977–981. doi:10.1271/bbb.65.977
Acknowledgements
The work of S.Z. and V.S.-S. is supported by the Austrian Science Fund (FWF): grants P20559 and T390 to V.S.-S..
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Hartl, L., Zach, S. & Seidl-Seiboth, V. Fungal chitinases: diversity, mechanistic properties and biotechnological potential. Appl Microbiol Biotechnol 93, 533–543 (2012). https://doi.org/10.1007/s00253-011-3723-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3723-3