Abstract
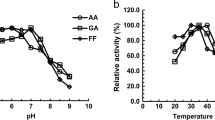
Acetaldehyde is a known mutagen and carcinogen. Active aldehyde dehydrogenase (ALDH) represents an important mechanism for acetaldehyde detoxification. A yeast strain XJ-2 isolated from grape samples was found to produce acetaldehyde dehydrogenase with a high activity of 2.28 U/mg and identified as Issatchenkia terricola. The enzyme activity was validated by oxidizing acetaldehyde to acetate with NAD+ as coenzyme based on the headspace gas chromatography analysis. A novel acetaldehyde dehydrogenase gene (ist-ALD) was cloned by combining SiteFinding-PCR and self-formed adaptor PCR. The ist-ALD gene comprised an open reading frame of 1,578 bp and encoded a protein of 525 amino acids. The predicted protein of ist-ALD showed the highest identity (73%) to ALDH from Pichia angusta. The ist-ALD gene was expressed in Escherichia coli, and the gene product (ist-ALDH) presented a productivity of 442.3 U/mL cells. The purified ist-ALDH was a homotetramer of 232 kDa consisting of 57 kDa-subunit according to the SDS-PAGE and native PAGE analysis. Ist-ALDH exhibited the optimal activity at pH 9.0 and 40°C, respectively. The activity of ist-ALDH was enhanced by K+, NH4+, dithiothreitol, and 2-mercaptoethanol but strongly inhibited by Ag+, Hg2+, Cu2+, and phenylmethyl sulfonylfluoride. In the presence of NAD+, ist-ALDH could oxidize many aliphatic, aromatic, and heterocyclic aldehydes, preferably acetaldehyde. Kinetic study revealed that ist-ALDH had a k cat value of 27.71/s and a k cat/K m value of 26.80 × 103/(mol s) on acetaldehyde, demonstrating ist-ALDH, a catalytically active enzyme by comparing with other ALDHs. These studies indicated that ist-ALDH was a potential enzymatic product for acetaldehyde detoxification.




Similar content being viewed by others
References
Abriola DP, MacKerell AD, Pietruszko R (1990) Correlation of loss of activity of human aldehyde dehydrogenase with reaction of bromoacetophenone with glutamic acid-268 and cysteine-302 residues. Partial-sites reactivity of aldehyde dehydrogenase. Biochem J 266(1):179–187
Ahvazi B, Coulombe R, Delarge M, Vedadi M, Zhang L, Meighen E, Vrielink A (2000) Crystal structure of the NADP+-dependent aldehyde dehydrogenase from Vibrio harveyi: structural implications for cofactor specificity and affinity. Biochem J 349:853–861
Bostian KA, Betts GF (1978) Rapid purification and properties of potassium-activated aldehyde dehydrogenase from Saccharomyces cerevisiae. Biochem J 173:773–786
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Brethauer S, Wyman CE (2010) Review: continuous hydrolysis and fermentation for cellulosic ethanol production. Bioresour Technol 101:4862–4874
Davis BJ (1964) Disc electrophoresis-II method and application to human serum proteins. Ann NY Acad Sci 121:404–427
Dickinson FM (1996) The purification and some properties of the Mg2+-activated cytosolic aldehyde dehydrogenase of Saccharomyces cerevisiae. Biochem J 315:393–399
Ho KK, Weiner H (2005) Isolation and characterization of an aldehyde dehydrogenase encoded by the aldB gene of Escherichia coli. J Bacteriol 187:1067–1073
Homann N, Tillonen J, Meurman JH, Rintamaki H, Lindqvist C, Rautio M, Jousimies-Somer H, Salaspuro M (2000) Increased salivary acetaldehyde levels in heavy drinkers and smokers: a microbiological approach to oral cavity cancer. Carcinogenesis 21:663–668
International Agency for Research on Cancer (1999) Acetaldehyde. In: IARC monographs on the evaluation of carcinogenic risks to humans, vol. 71. International Agency for Research on Cancer, Lyon
Jo JE, Mohan Raj S, Rathnasingh C, Selvakumar E, Jung WC, Park S (2008) Cloning, expression, and characterization of an aldehyde dehydrogenase from Escherichia coli K-12 that utilizes 3-hydroxypropionaldehyde as a substrate. Appl Microbiol Biotechnol 81:51–60
Kurkivuori J, Salaspuro V, Kaihovaara P, Kari K, Rautemaa R, Gronroos L, Meurman JH, Salaspuro M (2007) Acetaldehyde production from ethanol by oral streptococci. Oral Oncol 43:181–186
Kurtzman CP, Fell JW (2000) The yeasts: a taxonomic study, 4th edn. Elsevier, Amsterdam
Kurtzman CP, Robnett CJ (1998) Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 73:331–371
Lachenmeier DW, Sohnius EM (2008) The role of acetaldehyde outside ethanol metabolism in the carcinogenicity of alcoholic beverages: evidence from a large chemical survey. Food Chem Toxicol 46:2903–2911
Lachenmeier DW, Kanteres F, Rehm J (2009) Carcinogenicity of acetaldehyde in alcoholic beverages: risk assessment outside ethanol metabolism. Addiction 104:533–550
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Lassen N, Estey T, Tanguay RL, Pappa A, Reimers MJ, Vasiliou V (2005) Molecular cloning, baculovirus expression, and tissue distribution of the zebrafish aldehyde dehydrogenase 2. Drug Metab Dispos 33:649–656
Li X, Li Y, Wei D, Li P, Wang L, Feng L (2010) Characterization of a broad-range aldehyde dehydrogenase involved in alkane degradation in Geobacillus thermodenitrificans NG80-2. Microbiol Res 165:706–712
Lo HF, Chen YJ (2010) Gene cloning and biochemical characterization of a NAD(P)+ -dependent aldehyde dehydrogenase from Bacillus licheniformis. Mol Biotechnol 46:157–167
Meaden PG, Dickinson FM, Mifsud A, Tessier W, Westwater J, Bussey H, Midgley M (1997) The ALD6 gene of Saccharomyces cerevisiae encodes a cytosolic, Mg2+-activated acetaldehyde dehydrogenase. Yeast 13:1319–1327
Navarro-Avino JP, Prasad R, Miralles VJ, Benito RM, Serrano R (1999) A proposal for nomenclature of aldehyde dehydrogenases in Saccharomyces cerevisiae and characterization of the stress-inducible ALD2 and ALD3 genes. Yeast 15:829–842
Ohta T, Tani A, Kimbara K, Kawai F (2005) A novel nicotinoprotein aldehyde dehydrogenase involved in polyethylene glycol degradation. Appl Microbiol Biotechnol 68:639–646
Okibe N, Amada K, Hirano S, Haruki M, Imanaka T, Morikawa M, Kanaya S (1999) Gene cloning and characterization of aldehyde dehydrogenase from a petroleum-degrading bacterium, strain HD-1. J Biosci Bioeng 88:7–11
Park SE, Koo HM, Park YK, Park SM, Park JC, Lee OK, Park YC, Seo JH (2011) Expression of aldehyde dehydrogenase 6 reduces inhibitory effect of furan derivatives on cell growth and ethanol production in Saccharomyces cerevisiae. Bioresour Technol 102:6033–6038
Perozich J, Nicholas H, Wang BC, Lindahl R, Hempel J (1999) Relationships within the aldehyde dehydrogenase extended family. Protein Sci 8:137–146
Perozich J, Kuo I, Lindahl R, Hempel J (2001) Coenzyme specificity in aldehyde dehydrogenase. Chem Biol Interact 130–132:115–124
Priefert H, Krüger N, Jendrossek D, Schmidt B, Steinbüchel A (1992) Identification and molecular characterization of the gene coding for acetaldehyde dehydrogenase II (acoD) of Alcaligenes eutrophus. J Bacteriol 174:899–907
Raj S, Rathnasingh C, Jung WC, Selvakumar E, Park S (2010) A Novel NAD+-dependent aldehyde dehydrogenase encoded by the puuC gene of Klebsiella pneumoniae DSM 2026 that utilizes 3-hydroxypropionaldehyde as a substrate. Biotechnol Bioproc E 15:131–138
Roise D, Theiler F, Horvath SJ, Tomich JM, Richards JH, Allison DS, Schatz G (1988) Amphiphilicity is essential for mitochondrial presequence function. EMBO J 7:649–653
Roitel O, Sergienko E, Branlant G (1999) Dimers generated from tetrameric phosphorylating glyceraldehyde-3-phosphate dehydrogenase from Bacillus stearothermophilus are inactive but exhibit cooperativity in NAD Binding. Biochemistry 38:16084–16091
Rose TM, Henikoff JG, Henikoff S (2003) CODEHOP (consensus-degenerate hybrid oligonucleotide primer) PCR primer design. Nucleic Acids Res 31:3763–3766
Salaspuro V, Hietala J, Kaihovaara P, Pihlajarinne L, Marvola M, Salaspuro M (2002) Removal of acetaldehyde from saliva by a slow-release buccal tablet of l-cysteine. Int J Cancer 97:361–364
Salaspuro VJ, Hietala JM, Marvola ML, Salaspuro MP (2006) Eliminating carcinogenic acetaldehyde by cysteine from saliva during smoking. Cancer Epidemiol Biomarkers Prev 15:146–149
Sambrook J, Russel D (2007) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Sophos NA, Vasiliou V (2003) Aldehyde dehydrogenase gene superfamily: the 2002 update. Chem Biol Interact 143–144:5–22
Sripo T, Phongdara A, Wanapu C, Caplan AB (2002) Screening and characterization of aldehyde dehydrogenase gene from Halomonas salina strain AS11. J Biotechnol 95:171–179
Tamaki N, Nakamura M, Kimura K, Hama T (1977) Purification and properties of aldehyde dehydrogenase from Saccharomyces cerevisiae. J Biochem 82:73–79
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tan G, Gao Y, Shi M, Zhang X, He S, Chen Z, An C (2005) SiteFinding-PCR: a simple and efficient PCR method for chromosome walking. Nucleic Acids Res 33:e122
Tessier WD, Meaden PG, Dickinson FM, Midgley M (1998) Identification and disruption of the gene encoding the K+-activated acetaldehyde dehydrogenase of Saccharomyces cerevisiae. FEMS Microbiol Lett 164:29–34
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Vasiliou V, Pappa A (2000) Polymorphisms of human aldehyde dehydrogenases. Consequences for drug metabolism and disease. Pharmacology 61:192–198
Wang X, Mann CJ, Bai Y, Ni L, Weiner H (1998) Molecular cloning, characterization, and potential roles of cytosolic and mitochondrial aldehyde dehydrogenases in ethanol metabolism in Saccharomyces cerevisiae. J Bacteriol 180:822–830
Wang S, He J, Cui Z, Li S (2007) Self-formed adaptor PCR: a simple and efficient method for chromosome walking. Appl Environ Microbiol 73:5048–5051
Wang MF, Han CL, Yin SJ (2009) Substrate specificity of human and yeast aldehyde dehydrogenases. Chem Biol Interact 178:36–39
Xu J, Johnson RC (1995) aldB, an RpoS-dependent gene in Escherichia coli encoding an aldehyde dehydrogenase that is repressed by Fis and activated by Crp. J Bacteriol 177(11):3166–3175
Yamanaka Y, Kazuoka T, Yoshida M, Yamanaka K, Oikawa T, Soda K (2002) Thermostable aldehyde dehydrogenase from psychrophile, Cytophaga sp. KUC-1: enzymological characteristics and functional properties. Biochem Biophys Res Commun 298:632–637
Yoshida A, Rzhetsky A, Hsu LC, Chang C (1998) Human aldehyde dehydrogenase gene family. Eur J Biochem 251:549–557
Zhao Y, Lei M, Wu Y, Wang C, Zhang Z, Deng F, Wang H (2009) Molecular cloning and expression of the complete DNA sequence encoding NAD+-dependent acetaldehyde dehydrogenase from Acinetobacter sp. strain HBS-2. Ann Microbiol 59:97–104
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yao, Z., Zhang, C., Lu, F. et al. Gene cloning, expression, and characterization of a novel acetaldehyde dehydrogenase from Issatchenkia terricola strain XJ-2. Appl Microbiol Biotechnol 93, 1999–2009 (2012). https://doi.org/10.1007/s00253-011-3541-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3541-7