Abstract
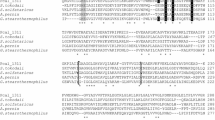
For the huge amount of chiral chemicals and precursors that can potentially be produced by biocatalysis, there is a tremendous need of enzymes with new substrate spectra, higher enantioselectivity, and increased activity. In this paper, a highly active alcohol dehydrogenase is presented isolated from Nocardia globerula that shows a unique substrate spectrum toward different prochiral aliphatic ketones and bulky ketoesters as well as thioesters. For example, the enzyme reduced ethyl 4-chloro-3-oxo butanoate with an ee >99% to (S)-4-chloro-3-hydroxy butanoate. Very interesting is also the fact that 3-oxobutanoic acid tert-butylthioester is reduced with 49.4% of the maximal activity while the corresponding tert-butyloxyester is not reduced at all. Furthermore, it has to be mentioned that acetophenone, a standard substrate for many known alcohol dehydrogenases, is not reduced by this enzyme. The enzyme was purified from wild-type N. globerula cells, and the corresponding 915-bp-long gene was determined, cloned, expressed in Escherichia coli, and applied in biotransformations. The N. globerula alcohol dehydrogenase is a tetramer of about 135 kDa in size as determined from gel filtration. Its sequence is related to several hypothetical 3-hydroxyacyl-CoA dehydrogenases whose sequences were derived by whole-genome sequencing from bacterial sources as well as known mammalian 3-hydroxyacyl-CoA dehydrogenases and ß-hydroxyacyl-CoA dehydrogenases from different clostridiae.



Similar content being viewed by others
References
Altschul SF, Madden TL, Schäffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Asanuma N, Kawato M, Ohkawara S, Hino T (2003) Characterization and transcription of the genes encoding enzymes involved in butyrate production in Butyrivibrio fibrisolvens. Curr Microbiol 47:203–207
Barycki JJ, O'Brien LK, Bratt JM, Zhang RG, Sanishvili R, Strauss AW, Banaszak LJ (1999) Biochemical characterization and crystal structure determination of human heart short chain L-3-hydroxyacyl-CoA dehydrogenase provide insights into catalytic mechanism. Biochemistry 38:5786–5798
Barycki JJ, O’Brien LK, Strauss AW, Banaszak LJ (2000) Sequestration of the active site by interdomain shifting—crystallographic and spectroscopic evidence for distinct conformations of L-3-hydroxyacyl-CoA dehydrogenase. J Biol Chem 275:27186–27196
Barycki JJ, O’Brien LK, Strauss AW, Banaszak LJ (2001) Glutamate 170 of human L-3-hydroxyacyl-CoA dehydrogenase is required for proper orientation of the catalytic histidine and structural integrity of the enzyme. J Biol Chem 276:36718–36726
Bayer M, Günther H, Simon H (1995) Purification and characterization of the NADH-dependent (S)-specific 3-oxobutyryl-CoA reductase from Clostridium tyrobutyricum. Arch Microbiol 163:310–312
Bennett GN, Rudolph FB (1995) The central metabolic pathway from acetyl-CoA to butyryl-Coa in Clostridium acetobutylicum. FEMS Microbiol Rev 17:241–249
Boynton ZL, Bennett GN, Rudolph FB (1996) Cloning, sequencing, and expression of clustered genes encoding beta-hydroxybutyryl-coenzyme A (CoA) dehydrogenase, crotonase, and butyryl-CoA dehydrogenase from Clostridium acetobutylicum ATCC 824. J Bacteriol 178:3015–3024
Bradford MM (1976) Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal Biochem 72:248–254
Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the clustal series of programs. Nucleic Acids Res 31:3497–3500
Colby GD, Chen JS (1992) Purification and properties of 3-hydroxybutyryl-coenzyme A dehydrogenase from Clostridium beijerinckii (“Clostridium butylicum”) NRRL B593. Appl Environ Microbiol 58:3297–3302
Ema T, Okita N, Ide S, Sakai T (2007) Highly enantioselective and efficient synthesis of methyl (R)-o-chloromandelate with recombinant E. coli: toward practical and green access to clopidogrel. Org Biomol Chem 5:1175–1176
Fiedler S, Steinbüchel A, Rehm BHA (2002) The role of the fatty acid beta-oxidation multienzyme complex from Pseudomonas oleovorans in polyhydroxyalkanoate biosynthesis: molecular characterization of the fadBA operon from P. oleovorans and of the enoyl-CoA hydratase genes phaJ from P. oleovorans and Pseudomonas putida. Arch Microbiol 178:149–160
Goldberg K, Schroer K, Lütz S, Liese A (2007) Biocatalytic ketone reduction—a powerful tool for the production of chiral alcohols—part I: processes with isolated enzymes. Appl Microbiol Biotechnol 76:237–248
Gröger H, Chamouleau F, Orologas N, Rollmann C, Drauz K, Hummel W, Weckbecker A, May O (2006) Enantioselective reduction of ketones with “Designer cells” at high substrate concentrations: highly efficient access to functionalized optically active alcohols. Angew Chem Int Ed 45:5677–5681
Hummel W (1997) New alcohol dehydrogenases for the synthesis of chiral compounds. Adv Biochem Eng/Biotechnol 58:145–184
Hummel W, Kula MR (1989) Dehydrogenases for the synthesis of chiral compounds. Eur J Biochem 184:1–13
Jörnvall H (1999) Multiplicity and complexity of SDR and MDR enzymes. Adv Exp Med Biol 463:359–364
Jörnvall H, Hoog JO, Persson B (1999) SDR and MDR: completed genome sequences show these protein families to be large, of old origin, and of complex nature. FEBS Lett 445:261–264
Jurček O, Wimmerová M, Wimmer Z (2008) Selected chiral alcohols: enzymic resolution and reduction of convenient substrates. Coord Chem Rev 252:767–781
Krozowski Z (1994) The short-chain alcohol dehydrogenase superfamily—variations on a common theme. J Steroid Biochem Mol Biol 51:125–130
Lampel KA, Uratani B, Chaudhry GR, Ramaley RF, Rudikoff S (1986) Characterization of the developmentally regulated Bacillus subtilis glucose dehydrogenase gene. J Bacteriol 166:238–243
Mullany P, Clayton CL, Pallen MJ, Slone R, al-Saleh A, Tabaqchali S (1994) Genes encoding homologues of three consecutive enzymes in the butyrate/butanol-producing pathway of Clostridium acetobutylicum are clustered on the Clostridium difficile chromosome. FEMS Microbiol Lett 124:61–67
Patel RN (2002) Microbial/enzymatic synthesis of chiral intermediates for pharmaceuticals. Enzyme Microb Technol 31:804–826
Patel RN (2003) Microbial/enzymatic synthesis of chiral pharmaceutical intermediates. Curr Opin Drug Discovery Dev 6:902–920
Patel RN (2008) Synthesis of chiral pharmaceutical intermediates by biocatalysis. Coord Chem Rev 252:659–701
Pollard DJ, Woodley JM (2007) Biocatalysis for pharmaceutical intermediates: the future is now. Trends Biotechnol 25:66–73
Reid MF, Fewson CA (1994) Molecular characterization of microbial alcohol dehydrogenases. Crit Rev Microbiol 20:13–56
Sanwal BD (1970) Regulatory characteristics of the diphosphopyridine nucleotide-specific malic enzyme of Escherichia coli. J Biol Chem 245:1212–1216
Schütte H, Flossdorf J, Sahm H, Kula MR (1976) Purification and properties of formaldehyde dehydrogenase and formate dehydrogenase from Candida boidinii. Eur J Biochem 62:151–160
Shimizu S, Ogawa J, Kataoka M, Kobayashi M (1997) Screening of novel microbial enzymes for the production of biologically and chemically useful compounds. Adv Biochem Eng/Biotechnol 58:45–87
Wolberg M, Villela M, Bode S, Geilenkirchen P, Feldmann R, Liese A, Hummel W, Müller M (2008) Chemoenzymatic synthesis of the chiral side-chain of statins: application of an alcohol dehydrogenase catalysed ketone reduction on a large scale. Bioprocess Biosyst Eng 31:183–191
Yang XY, Schulz H, Elzinga M, Yang SY (1991) Nucleotide sequence of the promoter and fadB gene of the fadBA operon and primary structure of the multifunctional fatty acid oxidation protein from Escherichia coli. Biochemistry 30:6788–6795
Youngleson JS, Jones DT, Woods DR (1989) Homology between hydroxybutyryl and hydroxyacyl coenzymeA dehydrogenase enzymes from Clostridium acetobutylicum fermentation and vertebrate fatty-acid beta-oxidation pathways. J Bacteriol 171:6800–6807
Acknowledgments
This work was supported by the “Bundesministerium für Bildung und Forschung” (Nachhaltige BioProduktion; Project “Entwicklung eines biokatalytischen und nachhaltigen Verfahrens zur industriellen Herstellung enantiomerenreiner Amine und Alkohole unter besonderer Berücksichtigung der Atomökonomie“).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Parkot, J., Gröger, H. & Hummel, W. Purification, cloning, and overexpression of an alcohol dehydrogenase from Nocardia globerula reducing aliphatic ketones and bulky ketoesters. Appl Microbiol Biotechnol 86, 1813–1820 (2010). https://doi.org/10.1007/s00253-009-2385-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-009-2385-x