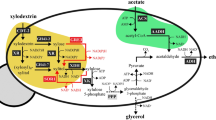
Abstract
Corynebacterium glutamicum R was metabolically engineered to broaden its sugar utilization range to d-xylose and d-cellobiose contained in lignocellulose hydrolysates. The resultant recombinants expressed Escherichia coli xylA and xylB genes, encoding d-xylose isomerase and xylulokinase, respectively, for d-xylose utilization and expressed C. glutamicum R bglF 317A and bglA genes, encoding phosphoenolpyruvate:carbohydrate phosphotransferase system (PTS) β-glucoside-specific enzyme IIBCA component and phospho-β-glucosidase, respectively, for d-cellobiose utilization. The genes were fused to the non-essential genomic regions distributed around the C. glutamicum R chromosome and were under the control of their respective constitutive promoter trc and tac that permitted their expression even in the presence of d-glucose. The enzyme activities of resulting recombinants increased with the increase in the number of respective integrated genes. Maximal sugar utilization was realized with strain X5C1 harboring five xylA–xylB clusters and one bglF 317A–bglA cluster. In both d-cellobiose and d-xylose utilization, the sugar consumption rates by genomic DNA-integrated strain were faster than those by plasmid-bearing strain, respectively. In mineral medium containing 40 g l−1d-glucose, 20 g l−1d-xylose, and 10 g l−1d-cellobiose, strain X5C1 simultaneously and completely consumed these sugars within 12 h and produced predominantly lactic and succinic acids under growth-arrested conditions.




Similar content being viewed by others
References
Aristidou A, Penttila M (2000) Metabolic engineering applications to renewable resource utilization. Curr Opin Biotechnol 11:187–198
Barrett E, Stanton C, Zelder O, Fitzgerald G, Ross RP (2004) Heterologous expression of lactose- and galactose-utilizing pathways from lactic acid bacteria in Corynebacterium glutamicum for production of lysine in whey. Appl Environ Microbiol 70:2861–2866
Dien BS, Cotta MA, Jeffries TW (2003) Bacteria engineered for fuel ethanol production: current status. Appl Microbiol Biotechnol 63:258–266
Fargione J, Hill J, Tilman D, Polasky S, Hawthorne P (2008) Land clearing and the biofuel carbon debt. Science 319:1235–1238
Gao Q, Zhang M, McMillan JD, Kompala DS (2002) Characterization of heterologous and native enzyme activity profiles in metabolically engineered Zymomonas mobilis strains during batch fermentation of glucose and xylose mixtures. Appl Biochem Biotechnol 98–100:341–355
Hahn-Hägerdal B, Karhumaa K, Fonseca C, Spencer-Martins I, Gorwa-Grauslund MF (2007) Towards industrial pentose-fermenting yeast strains. Appl Microbiol Biotechnol 74:937–953
Hernández-Montalvo V, Valle F, Bolivar F, Gosset G (2001) Characterization of sugar mixtures utilization by an Escherichia coli mutant devoid of the phosphotransferase system. Appl Microbiol Biotechnol 57:186–191
Inui M, Kawaguchi H, Murakami S, Vertès AA, Yukawa H (2004a) Metabolic engineering of Corynebacterium glutamicum for fuel ethanol production under oxygen-deprivation conditions. J Mol Microbiol Biotechnol 8:243–254
Inui M, Murakami S, Okino S, Kawaguchi H, Vertès AA, Yukawa H (2004b) Metabolic analysis of Corynebacterium glutamicum during lactate and succinate productions under oxygen deprivation conditions. J Mol Microbiol Biotechnol 7:182–196
Kawaguchi H, Vertès AA, Okino S, Inui M, Yukawa H (2006) Engineering of a xylose metabolic pathway in Corynebacterium glutamicum. Appl Environ Microbiol 72:3418–3428
Kawaguchi H, Sasaki M, Vertès AA, Inui M, Yukawa H (2008) Engineering of an l-arabinose metabolic pathway in Corynebacterium glutamicum. Appl Microbiol Biotechnol 77:1053–1062
Kinoshita S (1985) Glutamic acid bacteria. In: Demain AL, Solomon NA (eds) Biology of industrial microorganism. Bejamin/Cummings, London, pp 115–146
Kotrba P, Inui M, Yukawa H (2001) The ptsI gene encoding enzyme I of the phosphotransferase system of Corynebacterium glutamicum. Biochem Biophys Res Commun 289:1307–1313
Kotrba P, Inui M, Yukawa H (2003) A single V317A or V317M substitution in enzyme II of a newly identified beta-glucoside phosphotransferase and utilization system of Corynebacterium glutamicum R extends its specificity towards cellobiose. Microbiology 149:1569–1580
Mohagheghi A, Evans K, Chou YC, Zhang M (2002) Cofermentation of glucose, xylose, and arabinose by genomic DNA-integrated xylose/arabinose fermenting strain of Zymomonas mobilis AX101. Appl Biochem Biotechnol 98–100:885–898
Nichols NN, Dien BS, Bothast RJ (2001) Use of catabolite repression mutants for fermentation of sugar mixtures to ethanol. Appl Microbiol Biotechnol 56:120–125
Okino S, Suda M, Fujikura K, Inui M, Yukawa H (2008) Production of d-lactic acid by Corynebacterium glutamicum under oxygen deprivation. Appl Microbiol Biotechnol 78:449–454
Sakai S, Tsuchida Y, Nakamoto H, Okino S, Ichihashi O, Kawaguchi H, Watanabe T, Inui M, Yukawa H (2007) Effect of lignocellulose-derived inhibitors on growth of and ethanol production by growth-arrested Corynebacterium glutamicum R. Appl Environ Microbiol 73:2349–2353
Sambrook J, Fritsh EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. New York Cold Spring Harbor Laboratory Press, New York
Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A 74:5463–5467
Suzuki N, Okayama S, Nonaka H, Tsuge Y, Inui M, Yukawa H (2005) Large-scale engineering of the Corynebacterium glutamicum genome. Appl Environ Microbiol 71:3369–3372
Terasawa M, Yukawa H (1993) Industrial production of biochemicals by native immobilization. Bioprocess Technol 16:37–52
Vertès AA, Inui M, Kobayashi M, Kurusu Y, Yukawa H (1993) Presence of mrr- and mcr-like restriction systems in coryneform bacteria. Res Microbiol 144:181–185
Wood BE, Ingram LO (1992) Ethanol production from cellobiose, amorphous cellulose, and crystalline cellulose by recombinant Klebsiella oxytoca containing chromosomally integrated Zymomonas mobilis genes for ethanol production and plasmids expressing thermostable cellulase genes from Clostridium thermocellum. Appl Environ Microbiol 58:2103–2110
Yanase H, Nozaki K, Okamoto K (2005a) Ethanol production from cellulosic materials by genetically engineered Zymomonas mobilis. Biotechnol Lett 27:259–263
Yanase H, Yamamoto K, Sato D, Okamoto K (2005b) Ethanol production from cellobiose by Zymobacter palmae carrying the Ruminocuccus albus beta-glucosidase gene. J Biotechnol 118:35–43
Yukawa H, Omumasaba CA, Nonaka H, Kos P, Okai N, Suzuki N, Suda M, Tsuge Y, Watanabe J, Ikeda Y, Vertes AA, Inui M (2007) Comparative analysis of the Corynebacterium glutamicum group and complete genome sequence of strain R. Microbiology 153:1042–1058
Acknowledgments
We thank Crispinus A. Omumasaba (RITE) for helpful comments on the manuscript. This work was supported by a grant from the New Energy and Industrial Technology Development Organization (NEDO), Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sasaki, M., Jojima, T., Inui, M. et al. Simultaneous utilization of d-cellobiose, d-glucose, and d-xylose by recombinant Corynebacterium glutamicum under oxygen-deprived conditions. Appl Microbiol Biotechnol 81, 691–699 (2008). https://doi.org/10.1007/s00253-008-1703-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-008-1703-z