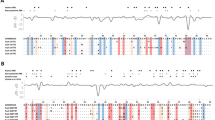
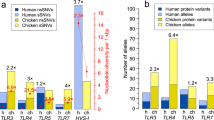
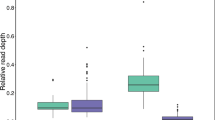
Abstract
Avian genomics, especially of non-model species, is in its infancy relative to mammalian genomics. Here, we describe the sequencing, assembly, and annotation of a new avian genome, that of the bananaquit Coereba flaveola (Passeriformes: Thraupidae). We produced ∼30-fold coverage of the genome with an assembly size of ca. 1.2 Gb, including approximately 16,500 annotated genes. Passerine birds, such as the bananaquit, are commonly infected by avian malarial parasites (Haemosporida), which presumably drive adaptive evolution of immunogenetic loci within the host genome. In the context of our research on the distribution of avian Haemosporida, we specifically characterized immune loci, including toll-like receptor (TLR) and major histocompatibility complex (MHC) genes. Additionally, we identified novel molecular markers in the form of single nucleotide polymorphisms (SNPs), both genome-wide and within identified immune loci. We discovered nine TLR genes and four MHC genes and identified five other TLR- or MHC- associated genes. Genome-wide, over 6 million high-quality SNPs were annotated, including 568 within TLR genes and 102 in MHC genes. This newly described genome and immune characterization expands the knowledge base for avian genomics and phylogenetics and allows for immune genotyping in the bananaquit, providing tools for the investigation of host-parasite coevolution.

Similar content being viewed by others
References
Alcaide M, Edwards SV (2011) Molecular evolution of the Toll-like receptor multigene family in birds. Molecular biology and evolution 28:1703-1715
Balakrishnan CN, Ekblom R, Volker M, Westerdahl H, Godinez R, Kotkiewicz H, Burt DW, Graves T, Griffin DK, Warren WC, Edwards SV (2010) Gene duplication and fragmentation in the zebra finch major histocompatibility complex. BMC Biol 8:29. doi:10.1186/1741-7007-8-29
Bellemain E, Bermingham E, Ricklefs RE (2008) The dynamic evolutionary history of the bananaquit (Coereba flaveola) in the Caribbean revealed by a multigene analysis. BMC Evol Biol 8:240. doi:10.1186/1471-2148-8-240
Benson G (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27:573–580
Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC (1987) The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature 329:512-518. doi:10.1038/329512a0
Boetzer M, Pirovano W (2012) Toward almost closed genomes with GapFiller. Genome Biol 13:R56. doi:10.1186/gb-2012-13-6-r56
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi:10.1093/bioinformatics/btu170
Bourlet Y, Behar G, Guillemot F, Frechin N, Billault A, Chausse AM, Zoorob R, Auffray C (1988) Isolation of Chicken Major Histocompatibility Complex Class Ii (B-L) Beta-Chain Sequences - Comparison with Mammalian Beta-Chains and Expression in Lymphoid Organs. Embo J 7:1031-1039
Brownlie R, Allan B (2011) Avian toll-like receptors. Cell Tissue Res 343:121–130. doi:10.1007/s00441-010-1026-0
Cantarel BL, Korf I, Robb SM, Parra G, Ross E, Moore B, Holt C, Sanchez Alvarado A, Yandell M (2008) MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res 18:188–196. doi:10.1101/gr.6743907
Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92. doi:10.4161/fly.19695
Cormican P, Lloyd AT, Downing T, Connell SJ, Bradley D, O’Farrelly C (2009) The avian Toll-like receptor pathway—subtle differences amidst general conformity. Dev Comp Immunol 33:967–973. doi:10.1016/j.dci.2009.04.001
DeWoody J, Abts K, Fahey A, Ji Y, Kimble S, Marra N, Wijayawardena B, Willoughby J (2013) Of contigs and quagmires: next-generation sequencing pitfalls associated with transcriptomic studies. Mol Ecol Resour 13:551–558. doi:10.1111/1755-0998.12107
Doležel J, Bartoš J, Voglmayr H, Greilhuber J (2003) Nuclear DNA content and genome size of trout and human. Cytometry A 51A:127–128. doi:10.1002/cyto.a.10013
Doyle JM, Katzner TE, Bloom PH, Ji Y, Wijayawardena BK, DeWoody JA (2014) The genome sequence of a widespread apex predator, the golden eagle (Aquila chrysaetos). PLoS One 9:e95599
Endo T, Ikeo K, Gojobori T (1996) Large-scale search for genes on which positive selection may operate. Mol Biol Evol 13:685–690
Fortin A, Stevenson MM, Gros P (2002) Susceptibility to malaria as a complex trait: big pressure from a tiny creature. Hum Mol Genet 11:2469–2478. doi:10.1093/hmg/11.20.2469
Fukui A, Inoue N, Matsumoto M, Nomura M, Yamada K, Matsuda Y, Toyoshima K, Seya T (2001) Molecular cloning and functional characterization of chicken toll-like receptors. A single chicken toll covers multiple molecular patterns. J Biol Chem 276:47143-47149. doi:10.1074/jbc.M103902200
Gallego C, Golenbock D, Gomez MA, Saravia NG (2011) Toll-like receptors participate in macrophage activation and intracellular control of Leishmania (Viannia) panamensis. Infect Immun 79:2871–2879
Gandon S, Michalakis Y (2002) Local adaptation, evolutionary potential and host–parasite coevolution: interactions between migration, mutation, population size and generation time. J Evol Biol 15:451–462. doi:10.1046/j.1420-9101.2002.00402.x
Gregory TR (2015) Animal Genome Size Database
Gremme G, Steinbiss S, Kurtz S (2013) GenomeTools: a comprehensive software library for efficient processing of structured genome annotations. IEEE/ACM Trans Comput Biol Bioinform 10:645–656. doi:10.1109/TCBB.2013.68
Griffiths R, Double MC, Orr K, Dawson RJG (1998) A DNA test to sex most birds. Mol Ecol 7:1071–1075. doi:10.1046/j.1365-294x.1998.00389.x
Grueber CE, Knafler GJ, King TM, Senior AM, Grosser S, Robertson B, Weston KA, Brekke P, Harris CLW, Jamieson IG (2015) Toll-like receptor diversity in 10 threatened bird species: relationship with microsatellite heterozygosity. Conserv Genet 16:595–611. doi:10.1007/s10592-014-0685-x
Gurevich A, Saveliev V, Vyahhi N, Tesler G (2013) QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi:10.1093/bioinformatics/btt086
Hess CM, Edwards SV (2002) The evolution of the major histocompatibility complex in birds: scaling up and taking a genomic approach to the major histocompatibility complex (MHC) of birds reveals surprising departures from generalities found in mammals in both large-scale structure and the mechanisms shaping the evolution of the MHC. Bioscience 52:423–431. doi:10.1641/0006-3568(2002)052[0423:teotmh]2.0.co;2
Hickson RE, Cann RL (1997) Mhc allelic diversity and modern human origins. J Mol Evol 45:589–598
Hill AV, Allsopp CE, Kwiatkowski D, Anstey NM, Twumasi P, Rowe PA, Bennett S, Brewster D, McMichael AJ, Greenwood BM (1991) Common west African HLA antigens are associated with protection from severe malaria. Nature 352:595–600
Holt C, Yandell M (2011) MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinf 12:491. doi:10.1186/1471-2105-12-491
Hughes AL (1992) Positive selection and interallelic recombination at the merozoite surface antigen-1 (MSA-1) locus of Plasmodium falciparum. Mol Biol Evol 9:381–393
Hughes M, Hughes A (1995) Natural selection on Plasmodium surface proteins. Mol Biochem Parasitol 71:99–113
Hughes CR, Miles S, Walbroehl JM (2008) Support for the minimal essential MHC hypothesis: a parrot with a single, highly polymorphic MHC class IIB gene. Immunogenetics 60:219–231. doi:10.1007/s00251-008-0287-1
Ji Y, DeWoody JA (2016) Genomic landscape of long terminal repeat retrotransposons (LTR-RTs) and solo LTRs as shaped by ectopic recombination in chicken and zebra finch. J Mol Evol 82:251–263. doi:10.1007/s00239-016-9741-0
Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong SY, Lopez R, Hunter S (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. doi:10.1093/bioinformatics/btu031
Kaufman J, Volk H, Wallny HJ (1995) A minimal-essential-Mhc and an unrecognized-Mhc—2 extremes in selection for polymorphism. Immunol Rev 143:63–88. doi:10.1111/j.1600-065X.1995.tb00670.x
Klein J, Bontrop RE, Dawkins RL, Erlich HA, Gyllensten UB, Heise ER, Jones PP, Parham P, Wakeland EK, Watkins DI (1990) Nomenclature for the major histocompatibility complexes of different species: a proposal. Immunogenetics 31:217-219
Kobe B, Kajava AV (2001) The leucine-rich repeat as a protein recognition motif. Curr Opin Struc Biol 11:725–732. doi:10.1016/S0959-440x(01)00266-4
Krishnegowda G, Hajjar AM, Zhu J, Douglass EJ, Uematsu S, Akira S, Woods AS, Gowda DC (2005) Induction of proinflammatory responses in macrophages by the glycosylphosphatidylinositols of Plasmodium falciparum: cell signaling receptors, glycosylphosphatidylinositol (gpi) structural requirement, and regulation of GPI activity. J Biol Chem 280:8606–8616
Kumar H, Kawai T, Akira S (2009) Toll-like receptors and innate immunity. Biochem Biophys Res Commun 388:621–625
Lamichhaney S, Berglund J, Almen MS, Maqbool K, Grabherr M, Martinez-Barrio A, Promerova M, Rubin CJ, Wang C, Zamani N, Grant BR, Grant PR, Webster MT, Andersson L (2015) Evolution of Darwin’s finches and their beaks revealed by genome sequencing. Nature 518:371–375. doi:10.1038/nature14181
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi:10.1093/bioinformatics/btp324
Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, Tang J, Wu G, Zhang H, Shi Y, Liu Y, Yu C, Wang B, Lu Y, Han C, Cheung DW, Yiu SM, Peng S, Xiaoqian Z, Liu G, Liao X, Li Y, Yang H, Wang J, Lam TW, Wang J (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18. doi:10.1186/2047-217X-1-18
MacDonald MR, Xia J, Smith AL, Magor KE (2008) The duck toll like receptor 7: genomic organization, expression and function. Mol Immunol 45:2055–2061. doi:10.1016/j.molimm.2007.10.018
Marcais G, Kingsford C (2011) A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27:764–770. doi:10.1093/bioinformatics/btr011
May RM, Anderson R (1983) Epidemiology and genetics in the coevolution of parasites and hosts. Proc R Soc Lond Ser B Biol Sci 219:281–313
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi:10.1101/gr.107524.110
Okazaki Y, Hume DA (2003) A guide to the mammalian genome—commentary. Genome Res 13:1267–1272. doi:10.1101/gr.1445603
Parra G, Bradnam K, Korf I (2007) CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23:1061–1067. doi:10.1093/bioinformatics/btm071
Philbin VJ, Iqbal M, Boyd Y, Goodchild MJ, Beal RK, Bumstead N, Young J, Smith AL (2005) Identification and characterization of a functional, alternatively spliced toll-like receptor 7 (TLR7) and genomic disruption of TLR8 in chickens. Immunology 114:507–521. doi:10.1111/j.1365-2567.2005.02125.x
Qi Y, Yan B, Chen S, Chen H, Wang M, Jia R, Zhu D, Liu M, Liu F, Yang Q, Sun K, Wu Y, Chen X, Jing B, Cheng A (2016) CpG oligodeoxynucleotide-specific goose TLR21 initiates an anti-viral immune response against NGVEV but not AIV strain H9N2 infection. Immunobiology 221:454-461. doi:10.1016/j.imbio.2015.11.005
Ricklefs RE, Fallon SM, Bermingham E (2004) Evolutionary relationships, cospeciation, and host switching in avian malaria parasites. Syst Biol 53:111–119
Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29:24–26. doi:10.1038/nbt.1754
Romanov MN, Tuttle EM, Houck ML, Modi WS, Chemnick LG, Korody ML, Mork EM, Otten CA, Renner T, Jones KC (2009) The value of avian genomics to the conservation of wildlife. BMC Genomics 10:S10
Sato A, Figueroa F, Mayer WE, Grant PR, Grant BR, Klein J (2000) Mhc class II genes of Darwin’s finches: divergence by point mutations and reciprocal recombination. Major Histocompatibility Complex:518–541
Schnoes AM, Brown SD, Dodevski I, Babbitt PC (2009) Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comput Biol 5:e1000605. doi:10.1371/journal.pcbi.1000605
Shapiro MD, Kronenberg Z, Li C, Domyan ET, Pan H, Campbell M, Tan H, Huff CD, Hu H, Vickrey AI, Nielsen SC, Stringham SA, Hu H, Willerslev E, Gilbert MT, Yandell M, Zhang G, Wang J (2013) Genomic diversity and evolution of the head crest in the rock pigeon. Science 339:1063-1067. doi:10.1126/science.1230422
Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I (2009) ABySS: a parallel assembler for short read sequence data. Genome Res 19:1117–1123. doi:10.1101/gr.089532.108
Smit AFA, Hubley R (2015) RepeatModeler Open-1.0 2008–2015
Smit AFA, Hubley R, Green P (2015) RepeatMasker Open-4.0 2013–2015. Institute for Systems Biology. http://repeatmasker.org
Stanke M, Waack S (2003) Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19(Suppl 2):ii215–ii225
Tommaso P, Moretti S, Xenarios I, Orobitg M, Montanyola A, Chang JM, Taly JF, Notredame C (2011) T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res 39:W13–W17. doi:10.1093/nar/gkr245
Valkiūnas G (2005) Avian malaria parasites and other haemosporidia. CRC Press, Boca Raton
Vicedomini R, Vezzi F, Scalabrin S, Arvestad L, Policriti A (2013) GAM-NGS: genomic assemblies merger for next generation sequencing. BMC Bioinf 14(Suppl 7):S6. doi:10.1186/1471-2105-14-S7-S6
Waltari E, Edwards SV (2002) Evolutionary dynamics of intron size, genome size, and physiological correlates in archosaurs. Am Nat 160:539–552. doi:10.1086/342079
Westerdahl H, Waldenström J, Hansson B, Hasselquist D, von Schantz T, Bensch S (2005) Associations between malaria and MHC genes in a migratory songbird. Proc R Soc B Biol Sci 272:1511–1518
Westerdahl H, Wittzell H, von Schantz T (2000) Mhc diversity in two passerine birds: no evidence far a minimal essential Mhc. Immunogenetics 52:92–100. doi:10.1007/s002510000256
Wicker T, Robertson JS, Schulze SR, Feltus FA, Magrini V, Morrison JA, Mardis ER, Wilson RK, Peterson DG, Paterson AH, Ivarie R (2005) The repetitive landscape of the chicken genome. Genome Res 15:126–136. doi:10.1101/gr.2438004
Wijayawardena BK, Minchella DJ, DeWoody JA (2013) Hosts, parasites, and horizontal gene transfer. Trends Parasitol 29:329–338. doi:10.1016/j.pt.2013.05.001
Wright NA, Gregory TR, Witt CC (2014) Metabolic ‘engines’ of flight drive genome size reduction in birds. Proc R Soc Lond B Biol Sci 281 doi:10.1098/rspb.2013.2780
Zhang G, Li C, Li Q, Li B, Larkin DM, Lee C, Storz JF, Antunes A, Greenwold MJ, Meredith RW, Odeen A, Cui J, Zhou Q, Xu L, Pan H, Wang Z, Jin L, Zhang P, Hu H, Yang W, Hu J, Xiao J, Yang Z, Liu Y, Xie Q, Yu H, Lian J, Wen P, Zhang F, Li H, Zeng Y, Xiong Z, Liu S, Zhou L, Huang Z, An N, Wang J, Zheng Q, Xiong Y, Wang G, Wang B, Wang J, Fan Y, da Fonseca RR, Alfaro-Nunez A, Schubert M, Orlando L, Mourier T, Howard JT, Ganapathy G, Pfenning A, Whitney O, Rivas MV, Hara E, Smith J, Farre M, Narayan J, Slavov G, Romanov MN, Borges R, Machado JP, Khan I, Springer MS, Gatesy J, Hoffmann FG, Opazo JC, Hastad O, Sawyer RH, Kim H, Kim KW, Kim HJ, Cho S, Li N, Huang Y, Bruford MW, Zhan X, Dixon A, Bertelsen MF, Derryberry E, Warren W, Wilson RK, Li S, Ray DA, Green RE, O’Brien SJ, Griffin D, Johnson WE, Haussler D, Ryder OA, Willerslev E, Graves GR, Alstrom P, Fjeldsa J, Mindell DP, Edwards SV, Braun EL, Rahbek C, Burt DW, Houde P, Zhang Y, Yang H, Wang J, Avian Genome C, Jarvis ED, Gilbert MT, Wang J (2014) Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346:1311–1320. doi:10.1126/science.1251385
Acknowledgements
We thank the Purdue Genomics Core Facility for the technical advice, Maria Pil from the Ricklefs Lab for the assistance with sample preparation, and members of the DeWoody Lab for the review of this manuscript. Funding was provided to JAD by Purdue through the University Faculty Scholar program and a seed grant from Purdue’s Department of Forestry and Natural Resources and to JA by a fellowship (P200A090324) in the area of Ecological and Environmental Engineering via the Department of Education’s Graduate Assistance in Area of National Need (GAANN) program.
Data management
The filtered sequencing reads, genome assembly, and annotated SNPs are available from NCBI (Bioproject ID: PRJNA353240). The genome annotation and sequences of transcripts and proteins can be found on Dryad (doi 10.5061/dryad.8182t).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(DOCX 254 kb)
Rights and permissions
About this article
Cite this article
Antonides, J., Ricklefs, R. & DeWoody, J.A. The genome sequence and insights into the immunogenetics of the bananaquit (Passeriformes: Coereba flaveola). Immunogenetics 69, 175–186 (2017). https://doi.org/10.1007/s00251-016-0960-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-016-0960-8