Abstract
Measuring individual-level heterozygosity in threatened species is one approach to understanding and mitigating losses of genetic diversity and the role of inbreeding depression in those populations. In many conservation contexts, this goal is approached by assaying levels of microsatellite diversity, and inference is often extended to functional genomic regions. Our study quantifies diversity of innate immunity toll-like receptor (TLR) genes in 10 threatened New Zealand birds across four avian orders, with an average of 20.1 individuals and 6.2 TLR loci (sequences averaging 850 bp in length) per species. We provide detailed TLR diversity statistics for these 10 species, which showed more evidence for genetic drift than balancing selection at TLR loci, with two possible exceptions (TLR1LA for hihi and TLR5 for kokako). Our observations also support a possible gene-duplication of TLR7 in rock wren, indicating that a TLR7 duplication previously observed in other passerines may have occurred early in the divergence of this order. In addition to these analyses of population-level TLR sequence diversity, we used an average of 14.6 polymorphic microsatellite loci per species to study, for the first time, the relationship between microsatellite internal relatedness (a measure of individual homozygosity) and TLR heterozygosity. There was no relationship between microsatellite and TLR heterozygosity of individuals within species, suggesting that the predictive power of microsatellites to evaluate functional diversity is poor, and highlighting the value of adding data from putatively functional genomic regions, such as TLRs, in the study of genetic diversity of threatened species. Overall this study provides valuable data for comparison with more widespread species, and facilitates research into the importance of TLR diversity in natural populations of conservation concern.


Similar content being viewed by others
References
Acevedo-Whitehouse K, Cunningham AA (2006) Is MHC enough for understanding wildlife immunogenetics? Trends Ecol Evol 21:433–438
Alcaide M, Edwards SV (2011) Molecular evolution of the toll-like receptor multigene family in birds. Mol Biol Evol 28:1703–1715
Alho JS, Lillandt BG, Jaari S, Merila J (2009) Multilocus heterozygosity and inbreeding in the Siberian jay. Conserv Genet 10:605–609. doi:10.1007/s10592-008-9588-z
Alho JS, Välimäki K, Merilä J (2010) Rhh: an R extension for estimating multilocus heterozygosity and heterozygosity–heterozygosity correlation. Mol Ecol Resour 10:720–722. doi:10.1111/j.1755-0998.2010.02830.x
Allendorf FW, Luikart G (2007) Conservation and the genetics of populations. Blackwell, Oxford
Amos W, Worthington Wilmer J, Fullard K et al (2001) The influence of parental relatedness on reproductive success. Proc R Soc Lond Ser B 268:2021–2027
Areal H, Abrantes J, Esteves P (2011) Signatures of positive selection in Toll-like receptor (TLR) genes in mammals. BMC Evol Biol 11:368. doi:10.1186/1471-2148-11-368
Babik W, Taberlet P, Ejsmond MJ, Radwan J (2009) New generation sequencers as a tool for genotyping of highly polymorphic multilocus MHC system. Mol Ecol Resour 9:713–719. doi:10.1111/j.1755-0998.2009.02622.x
Bainová H, Králová T, Bryjová A et al (2014) First evidence of independent pseudogenization of Toll-like receptor 5 in passerine birds. Dev Comp Immunol 45:151–155
Balloux F, Amos W, Coulson T (2004) Does heterozygosity estimate inbreeding in real populations? Mol Ecol 13:3021–3031
Barreiro LB, Ben-Ali M, Quach H et al (2009) Evolutionary dynamics of human Toll-like receptors and their different contributions to host defense. PLoS Genet 5:e1000562. doi:10.1371/journal.pgen.1000562
Basse B, Flux I, Innes J (2003) Recovery and maintenance of North Island kokako (Callaeas cinerea wilsoni) populations through pulsed pest control. Biol Conserv 109:259–270. doi:10.1016/S0006-3207(02)00154-4
Bates D, Maechler M (2009) lme4: linear mixed-effects models using S4 classes
BirdLife International (2013) Cyanoramphus novaezelandiae. IUCN 2013 IUCN Red List Threat. Species Version 20132. BirdLife International, Cambridge
Boessenkool S, Taylor SS, Tepolt CK et al (2007) Large mainland populations of South Island robins retain greater genetic diversity than offshore island refuges. Conserv Genet 8:705–714
Bollmer JL, Dunn PO, Whittingham LA, Wimpee C (2010) Extensive MHC class II B gene duplication in a Passerine, the common yellowthroat (Geothlypis trichas). J Hered 101:448–460. doi:10.1093/jhered/esq018
Bollmer JL, Ruder EA, Johnson JA et al (2011) Drift and selection influence geographic variation at immune loci of prairie-chickens. Mol Ecol 20:4695–4706. doi:10.1111/j.1365-294X.2011.05319.x
Brekke P, Bennett PM, Santure AW, Ewen JG (2011) High genetic diversity in the remnant island population of hihi and the genetic consequences of re-introduction. Mol Ecol 20:29–45. doi:10.1111/j.1365-294X.2010.04923.x
Casquet J, Thebaud C, Gillespie RG (2012) Chelex without boiling, a rapid and easy technique to obtain stable amplifiable DNA from small amounts of ethanol-stored spiders. Mol Ecol Resour 12:136–141. doi:10.1111/j.1755-0998.2011.03073.x
Chapman JR, Nakagawa S, Coltman DW et al (2009) A quantitative review of heterozygosity–fitness correlations in animal populations. Mol Ecol 18:2746–2765. doi:10.1111/j.1365-294X.2009.04247.x
Clout MN (2006) A celebration of kakapo: progress in the conservation of an enigmatic parrot. Notornis 53:1–2
Coltman DW, Pilkington JG, Smith JA, Pemberton J (1999) Parasite-mediated selection against inbred Soay sheep in a free-living island population. Evolution 53:1259–1267
Cormican P, Lloyd AT, Downing T et al (2009) The avian toll-like receptor pathway—subtle differences amidst general conformity. Dev Comp Immunol 33:967–973. doi:10.1016/j.dci.2009.04.001
Dawe MR (1979) Behaviour and ecology of the red-crowned kakariki (Cyanoramphus novaezelandiae) in relation to management. Department of Zoology, University of Auckland, Auckland
Deakin JE (2012) Marsupial genome sequences: providing insight into evolution and disease. Scientifica. doi:10.6064/2012/543176
Duncan RP, Blackburn TM (2004) Extinction and endemism in the New Zealand avifauna. Glob Ecol Biogeogr 13:509–517
Ericson PGP, Christidis L, Cooper A et al (2002) A Gondwanan origin of passerine birds supported by DNA sequences of the endemic New Zealand wrens. Proc R Soc Lond B 269:235–241. doi:10.1098/rspb.2001.1877
Ewens WJ (1972) The sampling theory of selectively neutral alleles. Theor Popul Biol 3:87–112
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Galan M, Guivier E, Caraux G et al (2010) A 454 multiplex sequencing method for rapid and reliable genotyping of highly polymorphic genes in large-scale studies. BMC Genom 11:296
Garrigan D, Hedrick PW (2003) Perspective: detecting adaptive molecular polymorphism: lessons from the MHC. Evolution 57:1707–1722. doi:10.1111/j.0014-3820.2003.tb00580.x
Gaze PD (1985) Distribution of yellowheads (Mohoua ochrocephala). Notornis 32:261–269
Gelman A, Su Y-S, Yajima M, et al (2009) arm: data analysis using regression and multilevel/hierarchical models. R-package. Available at http://CRAN.R-project.org/package=arm
Grueber CE, Jamieson IG (2011) Low genetic diversity and small population size of Takahe Porphyrio hochstetteri upon European arrival in New Zealand. Ibis 153:384–394
Grueber CE, Jamieson IG (2013) Primers for amplification of innate immunity Toll-like receptors in threatened birds of the Apterygiformes, Gruiformes, Psittaciformes and Passeriformes. Conserv Genet Resour 5:1043–1047. doi:10.1007/s12686-013-9965-x
Grueber CE, King TM, Waters JM, Jamieson IG (2008a) Isolation and characterization of microsatellite loci from the endangered New Zealand takahe (Gruiformes; Rallidae; Porphyrio hochstetteri). Mol Ecol Resour 8:884–886. doi:10.1111/j.1755-0998.2008.02098.x
Grueber CE, Wallis GP, Jamieson IG (2008b) Heterozygosity–fitness correlations and their relevance to studies on inbreeding depression in threatened species. Mol Ecol 17:3978–3984. doi:10.1111/j.1365-294X.2008.03910.x
Grueber CE, Waters JM, Jamieson IG (2011) The imprecision of heterozygosity–fitness correlations hinders the detection of inbreeding and inbreeding depression in a threatened species. Mol Ecol 20:67–79
Grueber CE, Wallis GP, King T, Jamieson IG (2012) Variation at innate immunity Toll-like receptor genes in a bottlenecked population of a New Zealand robin. PLoS One 7:e45011. doi:10.1371/journal.pone.0045011
Grueber CE, Wallis GP, Jamieson IG (2013) Genetic drift outweighs natural selection at toll-like receptor (TLR) immunity loci in a reintroduced population of a threatened species. Mol Ecol 22:4470–4482. doi:10.1111/mec.12404
Grueber CE, Wallis GP, Jamieson IG (2014) Episodic positive selection in the evolution of avian Toll-like receptor innate immunity genes. PLoS One. doi:10.1371/journal.pone.0089632
Hadfield JD (2010) MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. J Stat Softw 33:1–22
Hartmann SA, Schaefer HM, Segelbacher G (2014) Genetic depletion at adaptive but not neutral loci in an endangered bird species. Mol Ecol. doi:10.1111/mec.12975
Hedrick PW (1998) Balancing selection and MHC. Genetica 104:207–214
Hedrick PW (2012) What is the evidence for heterozygote advantage selection? Trends Ecol Evol 27:698–704. doi:10.1016/j.tree.2012.08.012
Higgins PJ (1999) Handbook of Australian, New Zealand and Antarctic birds. Oxford University Press, Melbourne
Higgins PJ, Peter JM (2002) Handbook of Australian, New Zealand and Antarctic birds. Volume 6: Pardalotes to shrike-thrushes. Oxford University Press, Melbourne
Higham T, Anderson A, Jacomb C (1999) Dating the first New Zealanders: the chronology of Wairau Bar. Antiquity 73:420–427
Holm S (1979) A simple sequentially rejective multiple test procedure. Scand J Stat 6:65–70
Holzapfel S, Robertson HA, McLennan JA et al (2009) Kiwi (Apteryx spp.) recovery plan, 2008–2018. Department of Conservation, Wellington
Hoosen S, Jamieson IG (2003) The distribution and current status of New Zealand saddleback Philesturnus carunculatus. Bird Conserv Int 13:79–95
Hudson QJ, Wilkins JR, Waas JR, Hogg ID (2000) Low genetic variability in small populations of New Zealand kokako Callaeas cinera wilsoni. Biol Conserv 96:105–112
Innes J, Hay R, Flux I et al (1999) Successful recovery of North Island kokako Callaeas cinerea wilsoni populations, by adaptive management. Biol Conserv 87:201–214. doi:10.1016/S0006-3207(98)00053-6
Jamieson IG, Tracy LN, Fletcher D, Armstrong DP (2007) Moderate inbreeding depression in a reintroduced population of North Island robins. Anim Conserv 10:95–102. doi:10.1111/j.1469-1795.2006.00078.x
Jepson A, Banya W, Sisay-Joof F et al (1997) Quantification of the relative contribution of major histocompatibility complex (MHC) and non-MHC genes to human immune responses to foreign antigens. Infect Immun 65:872–876
Keestra AM, de Zoete MR, Bouwman LI et al (2013) Unique features of chicken Toll-like receptors. Dev Comp Immunol 41:316–323. doi:10.1016/j.dci.2013.04.009
Keller LF, Waller DM (2002) Inbreeding effects in wild populations. Trends Ecol Evol 17:230–241. doi:10.1016/S0169-5347(02)02489-8
Kirk H, Freeland JR (2011) Applications and implications of neutral versus non-neutral markers in molecular ecology. Int J Mol Sci 12:3966–3988
Lee WG, Jamieson IG (2001) The takahe: fifty years of conservation management and research. University of Otago Press, Dunedin
Lettink M, Jamieson IG, Millar CD, Lambert DM (2002) Mating system and genetic variation in the endangered New Zealand takahe. Conserv Genet 3:427–434
Levene H (1949) On a matching problem arising in genetics. Ann Math Stat 20:91–94. doi:10.1214/aoms/1177730093
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. doi:10.1093/bioinformatics/btp187
Ljungqvist M, Akesson M, Hansson B et al (2010) Do microsatellites reflect genome-wide genetic diversity in natural populations? A comment on Väli et al. (2008). Mol Ecol 19:851–855
Meglécz E, Piry S, Desmarais E et al (2011) SESAME (SEquence Sorter & AMplicon Explorer): genotyping based on high-throughput multiplex amplicon sequencing. Bioinformatics 27:277–278. doi:10.1093/bioinformatics/btq641
Merton DV (1975) The saddleback: its status and conservation. In: Martin RD (ed) Breeding endangered species in captivity. Academic Press, London, pp 61–74
Michelsen-Heath S, Gaze P (2007) Changes in abundance and distribution of the rock wren (Xenicus gilviventris) in the South Island, New Zealand. Notornis 54:71–78
Miller HC, Lambert DM (2004) Genetic drift outweighs balancing selection in shaping post-bottleneck major histocompatibility complex variation in New Zealand robins (Petroicidae). Mol Ecol 13:3709–3721
Miller JM, Malenfant RM, David P et al (2014) Estimating genome-wide heterozygosity: effects of demographic history and marker type. Heredity 112:240–247. doi:10.1038/hdy.2013.99
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
O’Donnell CFJ, Roberts A, Lyall J (2002) Mohua (yellowhead) recovery plan 2002–2012. Department of Conservation, Wellington
Piertney SB, Oliver MK (2006) The evolutionary ecology of the major histocompatibility complex. Heredity 96:7–21. doi:10.1038/sj.hdy.6800724
Powlesland RG, Merton DV, Cockrem JF (2006) A parrot apart: the natural history of the kakapo (Strigops habroptilus), and the context of its conservation management. Notornis 53:3–26
R Core Team (2013) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Robertson BC, Minot EO, Lambert DM (2000) Microsatellite primers for the kakapo (Strigops habroptilus) and their utility in other parrots. Conserv Genet 1:93–95
Seutin G, White BN, Boag PT (1991) Preservation of avian blood and tissue samples for DNA analyses. Can J Zool 69:82–90
Slate J, David P, Dodds KG et al (2004) Understanding the relationship between the inbreeding coefficient and multilocus heterozygosity: theoretical expectations and empirical data. Heredity 93:255–265
Spurgin LG, Richardson DS (2010) How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc R Soc B Biol Sci 277:979–988
Szulkin M, Bierne N, David P (2010) Heterozygosity–fitness correlations: a time for reappraisal. Evolution 64:1202–1217
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Taylor SS, Jamieson IG (2008a) No evidence for loss of genetic variation following sequential translocations in extant populations of a genetically depauperate species. Mol Ecol 17:545–556. doi:10.1111/j.1365-294X.2007.03591.x
Taylor SS, Jamieson IG (2008b) No evidence for loss of genetic variation following sequential translocations in extant populations of a genetically depauperate species. Mol Ecol 17:545–556. doi:10.1111/j.1365-294X.2007.03591.x
Taylor C, Castro IC, Griffiths R (2005) Hihi/stitchbird (Notiomystis cincta) recovery plan, 2004–2009. Department of Conservation, Wellington
Taylor SS, Jamieson IG, Wallis GP (2007) Historic and contemporary levels of genetic variation in two New Zealand passerines with different histories of decline. J Evol Biol 20:2035–2047
Temperley N, Berlin S, Paton I et al (2008) Evolution of the chicken Toll-like receptor gene family: a story of gene gain and gene loss. BMC Genomics 9:62
Tracy L, Jamieson I (2011) Historic DNA reveals contemporary population structure results from anthropogenic effects, not pre-fragmentation patterns. Conserv Genet 12:517–526. doi:10.1007/s10592-010-0158-9
Turner AK, Begon M, Jackson JA et al (2011) Genetic diversity in cytokines associated with immune variation and resistance to multiple pathogens in a natural rodent population. PLoS Genet 7:e1002343
Uematsu S, Akira S (2008) Toll-like receptors (TLRs) and their ligands. Handb Exp Pharmacol 183:1–20. doi:10.1007/978-3-540-72167-3_1
Väli U, Einarsson A, Waits L, Ellegren H (2008) To what extent do microsatellite markers reflect genome-wide genetic diversity in natural populations? Mol Ecol 17:3808–3817. doi:10.1111/j.1365-294X.2008.03876.x
Veitch CR (2002) Eradication of Pacific rats (Rattus exulans) from Tiritiri Matangi Island, Hauraki Gulf, New Zealand. In: Veitch CR, Clout MN (eds) Turning the tide: the eradication of invasive species. IUCN, Gland, pp 360–364
Vinkler M, Albrecht T (2009) The question waiting to be asked: innate immunity receptors in the perspective of zoological research. Folia Zool 58:15–28
Walsh PS, Metzger DA, Higuchi R (1991) Chelex-100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 10:506–513
Watterson GA (1978) An analysis of multi-allelic data. Genetics 88:171–179
Werling D, Jann OC, Offord V et al (2009) Variation matters: TLR structure and species-specific pathogen recognition. Trends Immunol 30:124–130. doi:10.1016/j.it.2008.12.001
Westerdahl H (2007) Passerine MHC: genetic variation and disease resistance in the wild. J Ornithol 148:469–477. doi:10.1007/s10336-007-0230-5
Weston KA (2014) Conservation genetics of rock wren (Xenicus gilviventris). PhD, Department of Zoology, University of Otago
Weston KA, Robertson BC (2014) Isolation and characterisation of 14 microsatellites for the New Zealand rock wren Xenicus gilviventris. Conserv Genet Resour. doi:10.1007/s12686-013-0018-2
Wickes C, Crouchley D, Maxwell J (2009) Takahe recovery plan, 2007–2012. Department of Conservation, Wellington
Ziesemann B, Brunton DH, Castro IC (2011) Nesting success and breeding ecology in a high-density population of Brown Kiwi (Apteryx mantelli). Emu 111:148–154
Acknowledgments
We are grateful to those who generously provided samples and support for this study: Hugh Robertson, Oliver Overdyck, Tertia Thurley (New Zealand Department of Conservation); Kevin Parker (Massey University), John Ewen (Zoological Society of London) and Bethany Jackson (Auckland Zoo). We also thank Fiona Robertson for laboratory assistance and Ken Miller for preparation of Fig. 1. We are grateful for the continuing support of the New Zealand Department of Conservation, and in particular those Species Recovery Groups included in this study. This research was supported by the Allan Wilson Centre for Molecular Ecology and Evolution, the Marsden Fund, Landcare Research, University of Otago, a Royal Society Grant to PB and grants from Brian Mason Scientific and Technical Trust, Mohua Charitable Trust and JS Watson Conservation Trust to KW. CEG is currently supported by San Diego Zoo Global.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Appendix
Appendix
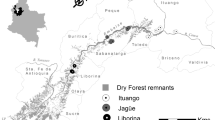
Here we provide species histories and details of study populations for the 10 threatened New Zealand native birds examined in this study. Polynesians were the first settlers to New Zealand from ~1200 AD (Higham et al. 1999), followed by a second colonisation by Europeans in the 1800s. Both immigration events brought about significant environmental changes, and much of New Zealand’s vertebrate fauna has been impacted. For example, New Zealand endemic birds have experienced dramatic population bottlenecks as a result of predation by introduced mammals (such as rats Rattus sp. and mustelids; New Zealand has no native terrestrial mammals except three species of bat), competition from introduced browsers (such as possums Trichosurus vulpecula), and habitat loss and fragmentation due to land conversion (Duncan and Blackburn 2004). These bottlenecks may have resulted in losses of genetic diversity, both at the population level (low numbers of alleles) and at the individual level (low observed heterozygosity). Sites mentioned herein are shown on the map in Fig. 1.
Apterygiformes: North Island brown kiwi Apteryx mantelli
The North Island brown kiwi is the most common of the five species of kiwi, currently numbering ~25,000, spread over four genetically distinct forms (Holzapfel et al. 2009). Despite intensive management, the species is classified under the New Zealand threat classification system as in “serious decline”, primarily as a result of human induced impacts (habitat loss and introduced mammalian predators, especially stoats Mustela erminea), and poor recruitment (Holzapfel et al. 2009; Ziesemann et al. 2011). We used feather samples from a population of brown kiwi near Purua, in the north of the North Island (currently numbers ~8,000; Holzapfel et al. 2009), collected as part of on-going management activities (H. Robertson pers. comm.).

Photo: Colin Miskelly, NZ Birds online (www.nzbirdsonline.org)
Gruiformes: takahe Porphyrio hochstetteri
Takahe were once widespread throughout the South Island of New Zealand, but are thought to have experienced dramatic declines since the arrival of Polynesian settlers in the 1600s and were even thought to be extinct by the 1900s (Lee and Jamieson 2001). A remnant population of birds was discovered in 1948 in the Murchison Mountains, Fiordland and has been subject to intense management since. The current population of <300 comprises birds in the original Murchison Mountains habitat, as well as translocated populations in several predator-free sanctuaries around the country (Wickes et al. 2009). We used samples from the source population, in Fiordland National Park, which were collected for a previous study (Lettink et al. 2002). Previous microsatellite analysis of takahe found very low levels of diversity, both in terms of numbers of polymorphic loci and numbers of alleles at variable loci (Grueber et al. 2008a), probably resulting from a prolonged population bottleneck (Grueber and Jamieson 2011).

Photo: Catherine Grueber
Psittaciformes: kakapo Strigops habroptilus
The kakapo is a large flightless bird, endemic to New Zealand and unique among parrots in its nocturnal, lek-breeding behaviours (Powlesland et al. 2006). The species is severely threatened by the introduction of stoats and rats and were the focus of possibly the earliest-known New Zealand conservation efforts, beginning in 1894. Sadly, only one Fiordland bird (“Richard Henry”) was ultimately rescued and translocated to Codfish Island along with members of a remnant population of birds from Stewart Island (Clout 2006). Very low levels of genetic diversity have been observed in kakapo with microsatellites (Robertson et al. 2000). All samples used in this study were collected as part of on-going management and are from Stewart Island-origin birds, i.e. we did not include samples from “Richard Henry” or his offspring. All extant kakapo (N ~124), except for Richard Henry’s offspring, descend from 62 Stewart Island birds (Powlesland et al. 2006).

Photo: Ian Jamieson
Psittaciformes: kakariki (red-crowned parakeet) Cyanoramphus novaezelandiae
Kakariki were once abundant on New Zealand’s North and South Islands until the introduction of predatory mammals and the destruction of suitable habitat significantly decreased their range (Higgins 1999). Kakariki are now considered effectively extinct from the mainland and only exist on Stewart Island and a number of offshore islands (BirdLife International 2013). We used samples collected from the wildlife sanctuary of Tiritiri Matangi in 2011 as part of concurrent research (B. Jackson pers. comm.). Since the introduction of approximately 90 birds to Tiritiri Matangi Island between 1974 and 1976 (Dawe 1979) and the subsequent eradication of kiore (Pacific rat Rattus exulans) from the island (Veitch 2002), the abundance of kakariki at the site has increased considerably.

Photo: Emily Weiser
Passeriformes: New Zealand rock wren Xenicus gilviventris
New Zealand rock wren are New Zealand’s only true alpine bird, living above the tree line for its entire life (Michelsen-Heath and Gaze 2007). Rock wren have been reported throughout much of the “main divide” of the Southern Alps, although a distribution study based on records of sightings over the past 100 years indicated that the species is declining (Michelsen-Heath and Gaze 2007). The samples we used here were collected from Fiordland, near the Homer Tunnel, as part of on-going research (Weston and Robertson 2014). This location is considered a stronghold for rock wren (Michelsen-Heath and Gaze 2007).

Photo: Bruce Robertson
Passeriformes: mohua (yellowhead) Mohoua ochrocephala
Mohua were formerly distributed across the entire South Island of New Zealand, but as a result of land conversion the species became fragmented into eight major forest patches (Gaze 1985; O’Donnell et al. 2002; Tracy and Jamieson 2011). Mohua are further threatened today by introduced mammalian predators, especially stoats (O’Donnell et al. 2002). Microsatellite-based studies of population structure and historical diversity (using museum specimens) revealed that the species has lost a significant amount of allelic diversity over the last 100 years and that a pattern of isolation by distance exists among contemporary mohua populations (Tracy and Jamieson 2011). We used samples from the Dart River Valley, Fiordland, for which DNA had been collected for a previous analysis (Tracy and Jamieson 2011); birds from this site are connected with those from other sites within a large forest patch.

Photo: Scott Mouat
Passeriformes: South Island robin Petroica australis australis
The South Island robin is a charismatic forest species endemic to New Zealand, belonging to the widespread Australasian family Petroicidae (Higgins and Peter 2002). Robins were previously distributed throughout the South Island, but the population has been increasingly fragmented as a result of habitat loss. Fragmentation is exacerbated by a reluctance of the birds to cross open water or unforested habitat, further isolating sub-populations of the species. A study of robin microsatellite diversity, in comparison to museum specimens, found little loss of diversity among contemporary birds as a whole, although birds from large mainland populations harboured more diversity than island birds (Boessenkool et al. 2007; Taylor et al. 2007). Here we use samples from one of the largest remnant robin populations, in the Eglinton River Valley, which were collected for a previous study (Boessenkool et al. 2007).

Photo: Ian Jamieson
Passeriformes: hihi (stitchbird) Notiomystis cincta
Once distributed throughout the North Island mainland and on northern offshore islands, hihi were extirpated from the mainland by the 1880s and persisted in a single remnant population on Little Barrier Island (Taylor et al. 2005). The Little Barrier Island population has been used as a source for subsequent translocations to establish populations on additional offshore islands (Taylor et al. 2005; Brekke et al. 2011). Microsatellite data revealed that the Little Barrier Island population has relatively high genetic diversity, possibly as a result of a high degree of extra-pair paternity in the species, reducing male reproductive variance (Brekke et al. 2011). We used samples from Little Barrier Island that were collected as part of on-going research (Brekke et al. 2011).

Photo: Paul Gibson
Passeriformes: North Island kokako Callaeas wilsoni
Kokako were formerly distributed across the North Island, but as a result of land conversion have become restricted to ~15 isolated forest fragments and introduced to several islands (Innes et al. 1999). The birds are particularly vulnerable to ship rats (Rattus rattus) and brushtail possums (Trichosurus vulpecula), so the recovery of kokako depends on the management of these invasive species (Basse et al. 2003). Microsatellite data have shown that the three largest kokako populations show only low levels of genetic differentiation (Hudson et al. 2000). Our samples were collected from the Mapara population, as part of ongoing management (O. Overdyck and T. Thurley, pers. comm.). Although the Mapara population is known to have declined to a small number of breeders in the 1990s, its recovery was rapid (Hudson et al. 2000).

Photo: Emily Weiser
Passeriformes: South Island saddleback Philesturnus carunculatus rufusator
South Island saddleback underwent an extreme population bottleneck when they were extirpated from the South Island of New Zealand at the time of human settlement and as a result of predation by invasive rats. Saddlebacks suffered a further severe bottleneck in the 1960s when the last known population, on Big South Cape Island, was reduced from around 1,000 birds to just 36 after rats arrived on the island. In 1964, these remaining birds were then moved to two rat-free sites, Big and Kaimohu Islands—the first threatened species translocation carried out by the New Zealand government through its Wildlife Service (later renamed the Department of Conservation) (Merton 1975). These populations grew and have been the source for several subsequent saddleback populations; the total population now numbers around 1,200 (Hoosen and Jamieson 2003). We used samples collected from Big Island as part of a previous study (Taylor and Jamieson 2008a, b).

Photo: Scott Mouat
Rights and permissions
About this article
Cite this article
Grueber, C.E., Knafler, G.J., King, T.M. et al. Toll-like receptor diversity in 10 threatened bird species: relationship with microsatellite heterozygosity. Conserv Genet 16, 595–611 (2015). https://doi.org/10.1007/s10592-014-0685-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-014-0685-x