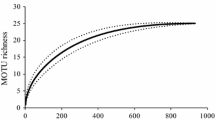
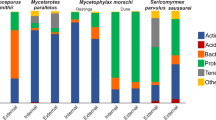
Abstract
Ant–plant mutualisms are conspicuous and ecologically important components of tropical ecosystems that remain largely unexplored in terms of insect-associated microbial communities. Recent work has revealed that ants in some ant–plant systems cultivate fungi (Chaetothyriales) within their domatia, which are fed to larvae. Using Pseudomyrmex penetrator/Tachigali sp. from French Guiana and Petalomyrmex phylax/Leonardoxa africana and Crematogaster margaritae/Keetia hispida, both from Cameroon, as models, we tested the hypothesis that ant–plant–fungus mutualisms co-occur with culturable Actinobacteria. Using selective media, we isolated 861 putative Actinobacteria from the three systems. All C. margaritae/K. hispida samples had culturable Actinobacteria with a mean of 10.0 colony forming units (CFUs) per sample, while 26 % of P. penetrator/Tachigali samples (mean CFUs 1.3) and 67 % of P. phylax/L. africana samples (mean CFUs 3.6) yielded Actinobacteria. The largest number of CFUs was obtained from P. penetrator workers, P. phylax alates, and C. margaritae pupae. 16S rRNA gene sequencing and phylogenetic analysis revealed the presence of four main clades of Streptomyces and one clade of Nocardioides within these three ant–plant mutualisms. Streptomyces with antifungal properties were isolated from all three systems, suggesting that they could serve as protective symbionts, as found in other insects. In addition, a number of isolates from a clade of Streptomyces associated with P. phylax/L. africana and C. margaritae/K. hispida were capable of degrading cellulose, suggesting that Streptomyces in these systems may serve a nutritional role. Repeated isolation of particular clades of Actinobacteria from two geographically distant locations supports these isolates as residents in ant–plant–fungi niches.




Similar content being viewed by others
References
Rico-Gray V, Oliveira PS (2007) The ecology and evolution of ant-plant interactions. The University of Chicago Press, Chicago, pp 1–331
Beattie A (1989) Myrmecotrophy: plants fed by ants. Trends Ecol Evol 4:172–176
Sagers CL, Ginger SM, Evans RD (2000) Carbon and nitrogen isotopes trace nutrient exchange in an ant-plant mutualism. Oecologia 123:582–586
Fischer RC, Wanek W, Richter A, Mayer V (2003) Do ants feed plants? A 15N labelling study of nitrogen fluxes from ants to plants in the mutualism of Pheidole and Piper. J Ecol 91:126–134
Janzen DH (1974) Epiphytic myrmecophytes in Sarawak: mutualism through the feeding of plants by ants. Biotropica 6:237–259
Rickson FR (1979) Absorption of animal tissue breakdown products into a plant stem-the feeding of a plant by ants. Am J Bot 66:87–90
Treseder KK, Davidson DW, Ehleringer JR (1995) Absorption of ant-provided carbon dioxide and nitrogen by a tropical epiphyte. Nature 375:137–139
Solano PJ, Dejean A (2004) Ant-fed plants: comparison between three geophytic myrmecophytes. Biol J Linn Soc 83:433–439
Watkins JE, Cardelús CL, Mack MC (2008) Ants mediate nitrogen relations of an epiphytic fern. New Phytol 180:5–8
Davidson DW, Mckey D (1993) The evolutionary ecology of symbiotic ant-plant relationships. J Hymenopt Res 2:13–83
Miehe H (1911) Untersuchungen über die javanische Myrmecodia. Abhandlungen der Math Klasse der Königlich-Sächsischen Gesellschaft der Wissenschaften 312–361
Defossez E, Selosse M-A, Dubois M-P et al (2009) Ant-plants and fungi: a new threeway symbiosis. New Phytol 182:942–949
Voglmayr H, Mayer V, Maschwitz U et al (2011) The diversity of ant-associated black yeasts: insights into a newly discovered world of symbiotic interactions. Fungal Biol 115:1077–1091
Blatrix R, Djiéto-Lordon C, Mondolot L et al (2012) Plant–ants use symbiotic fungi as a food source: new insight into the nutritional ecology of ant–plant interactions. Proc R Soc Biol Sci 279:3940–3947
Defossez E, Djiéto-Lordon C, McKey D et al (2010) Plant-ants feed their host plant, but above all a fungal symbiont to recycle nitrogen. Proc R Soc Biol Sci 278:1419–1426
Blatrix R, Bouamer S, Morand S, Selosee M-A (2009) Ant-plant mutualisms should be viewed as symbiotic communities. Plant Signal Behav 4:554–556
Lauth J, Ruiz-González MX, Orivel J (2011) New findings in insect fungiculture. Commun Integr Biol 4:728–730
Mueller UG, Gerardo NM, Aanen DK et al (2005) The evolution of agriculture in insects. Annu Rev Ecol Evol Syst 36:563–595
Currie CR, Mueller UG, Malloch D (1999) The agricultural pathology of ant fungus gardens. Proc Natl Acad Sci 96:7998–8002
Currie CR (2001) Prevalence and impact of a virulent parasite on a tripartite mutualism. Oecologia 128:99–106
Currie CR, Scott JA, Summerbell RA, Malloch D (1999) Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature 398:701–704
Currie CR, Bot ANM, Boomsma JJ (2003) Experimental evidence of a tripartite mutualism: bacteria protect ant fungus gardens from specialized parasites. Oikos 101:91–102
Fernández-Marín H, Zimmerman JK, Rehner SA, Wcislo WT (2006) Active use of the metapleural glands by ants in controlling fungal infection. Proc R Soc Biol Sci 273:1689–1695
Currie CR, Stuart AE (2001) Weeding and grooming of pathogens in agriculture by ants. Proc R Soc Biol Sci 268:1033–1039
Visser AA, Nobre T, Currie CR et al (2012) Exploring the potential for Actinobacteria as defensive symbionts in fungus-growing termites. Microb Ecol 63:975–985
Kaltenpoth M, Schmitt T, Polidori C et al (2010) Symbiotic streptomycetes in antennal glands of the South American digger wasp genus Trachypus (Hymenoptera, Crabronidae). Physiol Entomol 35:196–200
Kaltenpoth M, Goettler W, Dale C et al (2006) “Candidatus Streptomyces philanthi”, an endosymbiotic streptomycete in the antennae of Philanthus digger wasps. Int J Syst Evol Microbiol 56:1403–1411
Kaltenpoth M, Yildirim E, Gürbüz MF et al (2012) Refining the roots of the beewolf-Streptomyces symbiosis: antennal symbionts in the rare genus Philanthinus (Hymenoptera, Crabronidae). Appl Environ Microbiol 78:822–827
Hulcr J, Adams AS, Raffa K et al (2011) Presence and diversity of Streptomyces in Dendroctonus and sympatric bark beetle galleries across North America. Microb Ecol 61:759–768
Grubbs KJ, Biedermann PHW, Suen G et al (2011) Genome sequence of Streptomyces griseus strain XyelbKG-1, an ambrosia beetle-associated Actinomycete. J Bacteriol 193:2890–2891
Seipke RF, Barke J, Ruiz-Gonzalez MX et al (2012) Fungus-growing Allomerus ants are associated with antibiotic-producing actinobacteria. Anton Leeuw J Microb 101:443–447
Adams AS, Jordan MS, Adams SM et al (2011) Cellulose-degrading bacteria associated with the invasive woodwasp Sirex noctilio. ISME J 5:1323–1331
Poulsen M, Oh D-C, Clardy J, Currie CR (2011) Chemical analyses of wasp-associated Streptomyces bacteria reveal a prolific potential for natural products discovery. PLoS One 6:e16763
Miao V, Davies J (2010) Actinobacteria: the good, the bad, and the ugly. Anton Leeuw Int J G 98:143–150
Vasanthakumar A, Handelsman J, Schloss PD et al (2008) Gut microbiota of an invasive subcortical beetle, Agrilus planipennis Fairmaire, across various life stages. Environ Entomol 37:1344–1353
Pasti MB, Pometto AL III, Nuti MP, Crawford DL (1990) Lignin-solubilizing ability of Actinomycetes isolated from termite (Termitidae) Gut. Appl Environ Microbiol 56:2213–2218
Pasti MB, Belli ML (1985) Cellulolytic activity of Actinomycetes isolated from termites (Termitidae) gut. FEMS Microbiol Lett 26:107–112
Salem H, Kreutzer E, Sudakaran S, Kaltenpoth M (2013) Actinobacteria as essential symbionts in firebugs and cotton stainers (Hemiptera, Pyrrhocoridae). Environ Microbiol 15:1956–1968
Hsu SC, Lockwood JL (1975) Powdered chitin agar as a selective medium for enumeration of actinomycetes in water and soil. Appl Microbiol 29:422–426
Hillis DM, Marble BK, Larson A et al (1996) Nucleic acids IV: sequencing and cloning. In: Hillis DM, Moritz C, Mable BK (eds) Molecular systematics, 2nd edn. Sinauer Associates, Inc, Sunderland, pp 321–381
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acids techniques in bacterial systematics. Wiley, New York, pp 115–175
Cole JR, Wang Q, Cardenas E et al (2009) The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145
Scott JJ, Oh D, Yuceer MC et al (2008) Bacterial protection of beetle-fungus mutualism. Science 322:63
Cafaro MJ, Currie CR (2005) Phylogenetic analysis of mutualistic filamentous bacteria associated with fungus-growing ants. Can J Microbiol 51:441–446
Benson DA, Cavanaugh M, Clark K et al (2013) GenBank. Nucleic Acids Res 41:D36–D42
Pruitt KD, Tatusova T, Brown GR, Maglott DR (2012) NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res 40:D130–D135
Gao B, Gupta RS (2012) Phylogenetic framework and molecular signatures for the main clades of the phylum Actinobacteria. Microbiol Mol Biol Rev 76:66–112
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690
Stamatakis A, Hoover P, Rougemont J (2008) A rapid bootstrap algorithm for the RAxML web servers. Syst Biol 57:758–771
Letunic I, Bork P (2007) Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128
Letunic I, Bork P (2011) Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39:W475–W478
Cafaro MJ, Poulsen M, Little AEF et al (2011) Specificity in the symbiotic association between fungus-growing ants and protective Pseudonocardia bacteria. Proc R Soc Biol Sci 278:1814–1822
Poulsen M, Cafaro MJ, Erhardt DP et al (2009) Variation in Pseudonocardia antibiotic defence helps govern parasite-induced morbidity in Acromyrmex leaf-cutting ants. Environ Microbiol Rep 2:534–540
Takasuka TE, Book AJ, Lewin GR et al (2013) Aerobic deconstruction of cellulosic biomass by an insect-associated Streptomyces. Sci Rep 3:1–10
Hain T, Ward-Rainey N, Kroppenstedt RM et al (1997) Discrimination of Streptomyces albidoflavus strains based on the size and number of 16S-23S ribosomal DNA intergenic spacers. Int J Syst Bacteriol 47:202–206
Guo Y, Zheng W, Rong X, Huang Y (2008) A multilocus phylogeny of the Streptomyces griseus 16S rRNA gene clade: use of multilocus sequence analysis for streptomycete systematics. Int J Syst Evol Microbiol 58:149–159
Kaltenpoth M, Go W, Herzner G et al (2005) Symbiotic bacteria protect wasp larvae from fungal infestation. Curr Biol 15:475–479
Kaltenpoth M (2009) Actinobacteria as mutualists: general healthcare for insects? Trends Microbiol 17:529–535
Acknowledgments
We thank Erik Houck, Karen Bednar, Diana Xiao, and Dan Phillips for their assistance with lab work, and Gina Lewin and Adam Book for assistance in setting up the cellulose degradation assays. We thank Heidi Horn, Jonathan Klassen, and Charles Mason for technical suggestions and comments on this manuscript. ASH was partially funded by National Institutes of Health T32 GM07215-37. CRC received funding from National Science Foundation MCB-0702025 and DOE BER Office of Science DE-FC02-07ER64494.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Fig. 1
Representatives of bioassay evaluation of antifungal activity. Nineteen isolates were tested for their antifungal activity. a Trichoderma reesei control plate after complete growth over the full plate. b Nocardiodies sp. LaPpAH12 with no ZOI against T. reesei. c Streptomyces sp PsTaAH5 with a moderate ZOI against T. reesei. d Streptomyces sp KhCrAH316 with moderate ZOI against T. reesei. e Streptomyces sp PsTaAH124 with almost complete suppression of T. reesei (GIF 102 kb)
Supplemental Table 1
(XLSX 42 kb)
Rights and permissions
About this article
Cite this article
Hanshew, A.S., McDonald, B.R., Díaz Díaz, C. et al. Characterization of Actinobacteria Associated with Three Ant–Plant Mutualisms. Microb Ecol 69, 192–203 (2015). https://doi.org/10.1007/s00248-014-0469-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-014-0469-3