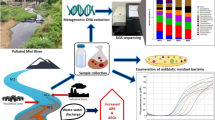
Abstract
Bacterial resistance to antibiotics has become a public health issue. Over the years, pathogenic organisms with resistance traits have been studied due to the threat they pose to human well-being. However, several studies raised awareness to the often disregarded importance of environmental bacteria as sources of resistance mechanisms. In this work, we analyze the diversity of antibiotic-resistant bacteria occurring in aquatic environments of the state of Rio de Janeiro, Brazil, that are subjected to distinct degrees of anthropogenic impacts. We access the diversity of aquatic bacteria capable of growing in increasing ampicillin concentrations through 16S rRNA gene libraries. This analysis is complemented by the characterization of antibiotic resistance profiles of isolates obtained from urban aquatic environments. We detect communities capable of tolerating antibiotic concentrations up to 600 times higher than the clinical levels. Among the resistant organisms are included potentially pathogenic species, some of them classified as multiresistant. Our results extend the knowledge of the diversity of antibiotic resistance among environmental microorganisms and provide evidence that the diversity of drug-resistant bacteria in aquatic habitats can be influenced by pollution.




Similar content being viewed by others
References
Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J (2010) Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol 8:251–259
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 5:403–410
Andersson DI, Levin BR (1999) The biological cost of antibiotic resistance. Curr Opin Microbiol 2:489–493
Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ (2006) New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl Environ Microbiol 72:5734–5741
Ball M, Gómez W, Magallanes X, Rosales R, Melfo A, Yarzábal L.A (2013) Bacteria recovered from high-altitude, tropical glacier in Venezuelan Andes. World J Microbol Biotechnol
Baquero F, Martínez JL, Cantón R (2008) Antibiotic and antibiotic resistance in water environments. Curr Opin Biotechnol 19:260–265
Bennett PM (2008) Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. Br J Pharmacol 153(Suppl 1):S347–S357
Bhullar K, Waglechner N, Pawlowski A, Koteva K, Banks E.D, Johnston M.D. et al. (2012) Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS ONE
Cantón R (2009) Antibiotic resistance genes from the environment: a perspective through newly identified antibiotic resistance mechanisms in the clinical setting. Clin Microbiol Infect 15(Suppl 1):20–25
Carlet J, Jarlier V, Harbarth S, Voss A, Gossens H, Pittet D (2012) Ready for a world without antibiotics? The Pensières antibiotic resistance call to action. Antimicrob Restist Infect Control 14:11–23
Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK, Lim YW (2007) EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol 57:2259–2261
Cole JR, Wang Q, Crdenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarell DM, Marsh T, Garrity GM, Tiedje JM (2009) The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:141–145
Czekalski N, Berthold T, Caucci S, Egli A, Bürgmann H (2012) Increased levels of multiresistant bacteria and resistance genes after wastewater treatment and their dissemination into Lake Geneva, Switzerland. Front Microbiol 3:106
D'Costa VM, Griffiths E, Wright GD (2007) Expanding the soil antibiotic resistome: exploring environmental diversity. Curr Opin Microbiol 10:481–489
Dantas G, Sommer MOA, Oluwasegun RD, Church GM (2008) Bacteria subsisting on antibiotics. Science 320:100–103
Davies J (2006) Are antibiotics naturally antibiotics? J Indust Microbiol Biotechnol 33:496–499
De Souza MJ, Nair S, Loka Bharathi PA, Chandramohan D (2006) Metal and antibiotic-resistance in psychrotrophic bacteria from Antarctic Marine waters. Ecotoxicology 15:379–384
Deris JB, Kim M, Zhang Z, Okano H, Hersmen R, Groisman A, Hwa T (2013) The innate growth bistability and fitness landscapes of antibiotic-resistant bacteria. Science 342:12374351–12374358
Dethlefsen, L., Huse, S, Sogin, M.L., Relman, D.A. (2008) The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biology.
D’Costa VM, King CE, Kalan L, Morar M, Sung WWL, Schwarz C et al (2011) Antibiotic resistance is ancient. Nature 477:457–461
D’Costa VM, McGrann KM, Hughes DW, Wright GD (2006) Sampling the antibiotic resistome. Science 311:374–377
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Edwing B, Hillier L, Wendl M, Green P (1998) Basecalling of automated sequencer traces using phred. I. Accuracy assessment. Gen Res 8:175–185
Forbserg KJ, Reyes A, Wang B, Selleck EM, Sommer MOA, Dantas G (2012) The shared antibiotic resistome of soil bacteria and human pathogens. Science 337:1107–1111
Gaze WH, Krone SM, Joakim-Larsson DG, Li XZ, Robinson JA, Simonet P et al (2013) Influence of humans on the evolution and mobilization of environmental antibiotic resistome. Emerg Infect Dis 19:1–7
Gifford DR, MacLean RC (2013) Evolutionary reversals of antibiotic resistance in experimental populations of Pseudomonas aeruginosa. Evolution 67:2973–2981
Gillings MR, Stokes HW (2012) Are humans increasing bacterial evolvability? Trends Ecol Evol 27:346–352
Goh E, Yim G, Tsui W, McClure J, Surette MG, Davies J (2002) Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. PNAS 99:17025–17030
Gregoracci G.B, Nascimento J.R, Cabral A.S, Paranhos R, Valentin J.L, Thomspon, C.C, Thompson F.L (2012) Structuring of bacterioplankton diversity in a large tropical bay. PLoS ONE
Höjgård S. Antibiotic resistance—why is the problem so difficult to solve? (2012) Infect Ecol Epidemol.
Jernberg C, Löfmark S, Edlund C, Jansson JK (2007) Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J 1:56–66
Juhas M (2013) Horizontal gene transfer in human pathogens. Crit Rev in Microbiol
Kristiansson E, Fick J, Janzon A, Grabic R, Rutgersson C, Weijdegår B, Söderström H, Larsson D.G (2011) Pyrosequencing of antibiotic-contaminated river sediments reveals high levels of resistance and gene transfer elements. PLoS ONE
Kümmerer K (2009) Antibiotics in the aquatic environment—a review. Part II. Chemosphere 75:435–441
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematic. Wiley, New York, pp 115–175
Levy SB, Marshall B (2004) Antibacterial resistance worldwide: causes, challenges and responses. Nat Med 10(12 Suppl):122–129
Linares JF, Gustafsson I, Baquero F, Martínez JL (2006) Antibiotics as intermicrobial signaling molecules instead of weapons. PNAS 103:19484–19489
Lozupone C, Hamady M, Knight R (2006) UniFrac - an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinforma 7:371–384
Lupo A, Coyne S, Berendonk TU (2012) Origin and evolution of antibiotic resistance: the commom mechanisms of emergence and spread in water bodies. Front Microbiol 3:18
Martínez JL (2008) Antibiotics and antibiotic resistance genes in natural environments. Science 321:365–367
Martínez JL (2009) Environmental pollution by antibiotics and antibiotic resistance determinants. Environ Pollut 157:2893–2902
Martínez JL (2012) Natural antibiotic resistance and contamination by antibiotic resistance determinants: the two ages in the evolution of resistance to antimicrobials. Front Microbiol 3:1
Martínez JL, Baquero F, Andersson DI (2007) Predicting antibiotic resistance. Nat Rev Microbiol 5:958–965
Pai V, Rao VI, Rao SP (2010) Prevalence and antimicrobial susceptibility pattern of methicillin-resistant Staphylococcus aureus [MRSA] isolates at a tertiary care hospital in Mangalore, South India. J Lab Phys 2:82–84
Pruden A, Pei R, Storteboom H, Carlson KH (2006) Antibiotic resistance genes as emerging contaminants: studies in Northern Colorado. Environ Sci Technol 40:7745–7750
Santoro DO, Romão CMCA, Clementino MM (2012) Decreased aztreonam susceptibility among Pseudomonas aeruginosa isolates from hospital effluent treatment system and clinical samples. Int J Environ Health Res 22:560–570
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Schwartz T, Kohnen W, Jansen B, Obst U (2003) Detection of antibiotic-resistant bacteria and their resistance genes in wastewater, surface water and drinking water biofilms. FEMS Microbiol Ecol 43:325–335
Segawa T, Takeuchi N, Rivera A, Yamada A, Yoshimura Y, Barcaza G et al (2013) Distribution of antibiotic resistance in glacier environments. Environ Microbiol Rep 5:127–134
Sejas LM, Silbert S, Reis AO, Sader HS (2003) Avaliação da qualidade dos discos com antimicrobianos para testes de disco-difusão disponíveis comercialmente no Brasil. Jornal Brasileiro de Patologia e Medicina Laboratorial 39:27–35
Sengupta S, Chattopadhyay MK, Grossart HP (2013) The multifaceted roles of antibiotics and antibiotic resistance in nature. Front Microbiol 4:47
Silveira CB, Vieira RP, Cardoso AM, Paranhos R, Albano RM, Martins OB (2011) Influence of salinity on bacterioplankton communities from the Brazilian rain forest to the coastal Atlantic Ocean. PloS One
Sommer MO, Dantas G, Church GM (2009) Functional characterizathion of the antibiotic resitance reservoir in the human microflora. Science 325:1128–1131
Tacão M, Correia A, Henriques I (2012) Resistance to broad-spectrum antibiotics in aquatic systems: anthropogenic activities modulate the dissemination of blaCTX-M-like genes. Appl Environ Microbiol 78:4134–4140
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thevenon F, Adatte T, Wildi W, Poté J (2012) Antibiotic resistant bacteria/genes dissemination in lacustrine sediments highly increased following cultural eutrophication of Lake Geneva (Switzerland). Chemosphere 86:468–476
Vieira RP, Gonzalez AM, Cardoso AM, Oliveira DN, Albano RM, Clementino MM, Martins OB, Paranhos R (2008) Relationships between bacterial diversity and environmental variables in a tropical marine environment, Rio de Janeiro. Environ Microbiol 10:189–199
Vieira RP, Clementino MM, Cardoso AM, Oliveira DN, Albano RM, Gonzalez AM et al (2007) Archaeal Communities in a Tropical Estuarine Ecosystem: Guanabara Bay, Brazil. Microb Ecol 54:460–462
Vignesh S, Muthukumar K, James RA (2012) (2012) Antibiotic resistant pathogens versus human impacts: a study from three eco-regions of the Chennai coast, southern India. Mar Pollut Bull 64:790–800
Walsh F (2013) Investigating antibiotic resistance in non-clinical environments. Front Microbiol 4:19
Weissburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Wise R (2002) Antimicrobial resistance: priorities for action. J Antimicrob Chemother 49:585–586
World Health Organization (WHO) (2000) WHO Annual report on infectious disease: overcoming antimicrobial resistance. World Health Organization: Geneva, Switzerland, 2000. http://www.who.int/infectious-disease-report/2000/
World Health Organization (WHO) (2012) Fact sheet N°194: antimicrobial resistance. http://www.who.int/mediacentre/factsheets/fs194/en/index.html
Wright GD (2007) The antibiotic resistome: the nexus of chemical genetic diversity. Nat Rev Microbiol 5:175–186
Wright GD (2010) Antibiotic resistance in the environment: a link to the clinic? Curr Opin Microbiol 13:589–594
Yim G, Wang H, Davies J (2007) Antibiotics as signaling molecules. Philos Trans R Soc Lond B Biol Sci 362:1195–1200
Zhang Q, Lambert G, Liao D, Kim H, Robin K, Tung CK et al (2011) Acceleration of emergence of bacterial antibiotic resistance in connected microenvironments. Science 333:1764–1767
Zhang XX, Zhang T, Fang HH (2009) Antibiotic resistance genes in water environment. Appl Microbiol Biotechnol 82:397–414
Salloto GRB, Cardoso AM, Coutinho FH, Pinto LH, Vieira RP, Chaia C, Lima JL et al (2012) Pollution impacts on bacterioplankton diversity in a tropical urban coastal lagoon system. PLoS ONE 7(11):e51175
Singh GB (2004) β-Lactams in the new millennium. Part I: monobactams and carbapenems. Mini Rev Med Chem 4:69–92
Rajpara N, Patel A, Tiwari N, Bahuguna J, Antony A, Choudhury I et al (2009) Int J Antimicrob Agents 34(3):220–225
Ahmed AM, Nakagawa T, Arakawa E, Ramamurthy T, Shinoda S, Shimamoto T (2004) New aminoglycoside acetyltransferase gene, aac(3)-Id, in a class 1 integron from a multiresistant strain of Vibrio fluvialis isolated from an infant aged 6 months. J Antimicrob Chemother 53(6):947–951
Bonomo RA, Szabo D (2006) Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin Infect Dis 43:S49–S56
Quinteira S, Ferreira H, Peixe L (2005) First Isolation of blaVIM-2 in an environmental isolate of Pseudomonas pseudoalcaligenes. Antimicrob Agents Chemother 49(5):2140–2141
Tremaroli V, Fedi S, Turner RJ, Ceri H, Zannoni D (2008) Pseudomonas pseudoalcaligenes KF707 upon bioWlm formation on a polystyrene surface acquire a strong antibiotic resistance with minor changes in their tolerance to metal cations and metalloid oxyanions. Arch Microbiol 190(1):29–39
Nazl SA, Tariq P (2005) Prevalence and antibiogram pattern of Pseudomonas species causing secondary infectious among patients of pulomnary tuberculosis. Int Chem Pharm Med J 2(2):231–237
Rice LB (2009) The clinical consequences of antimicrobial resistance. Curr Opin Microbiol 12(5):476–481
Depardieu F, Podglajen I, Leclercq R, Collatz E, Courvalin P (2007) Modes and modulation of antibiotic resistance gene expression. Clin Microbiol Rev 20(1):79–114
Acknowledgments
This study was financed by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Fundação Carlos Chagas Filho de Amparo a Pesquisa do Estado do Rio de Janeiro (FAPERJ). We acknowledge genome studies facilities Johanna Döbereiner. Special thanks to Joyce Lemos Lima for preparing sequencing reactions.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Coutinho, F.H., Silveira, C.B., Pinto, L.H. et al. Antibiotic Resistance is Widespread in Urban Aquatic Environments of Rio de Janeiro, Brazil. Microb Ecol 68, 441–452 (2014). https://doi.org/10.1007/s00248-014-0422-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-014-0422-5