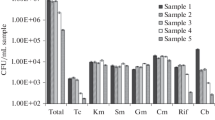
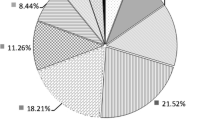
Abstract
The spread of antibiotic-resistance bacteria and antibiotic-resistance genes (ARGs) has been of concern worldwide. In this study, 114 Escherichia coli isolates were isolated from surface water samples of a lake to identify their susceptibility to antibiotics, including tetracycline (TC), gentamicin (GN), ampicillin (AMP), streptomycin (ST), oxytetracycline (OC), levofloxacin (LEV), nalidixic acid (NA), and sulfamethoxazole/trimethoprim (SFT). Isolates showing resistance to TC, GN, AMP, ST, OC, LEV, NA, and SFT occurred in 50, 76, 68, 71, 55, 32, 82, and 85 % of the total isolates, respectively. Thirty-seven different resistance patterns were identified, and the most abundant resistance profile (28 of 104) was TC/GN/AMP/ST/OC/LEV/NA/SFT. The occurrence of 29 ARGs were detected in their corresponding resistance clones, and 88 % of TC-resistance, 94 % of SFT-resistance, 90 % of AMP-resistance, 78 % of ST-resistance, and 72 % of quinolone-resistance clones can be described by their corresponding ARGs. It should be noted that most of these antibiotic-resistance clones harbored at least two corresponding ARGs, indicating that high frequencies of combined ARGs occurred in these isolates. In addition, 9 new types of DNA sequence of qnr(B) gene were obtained and were clustered into the same group as showed by phylogenetic trees analysis. These results suggest that the development of antibiotic resistance can be ascribed to the high frequency in the recombination of ARGs through horizontal gene transfer.



Similar content being viewed by others
References
Ahmed AM, Motoi Y, Sato M, Maruyama A, Watanabe H, Fukumoto Y et al (2007) Zoo animals as reservoirs of Gram-negative bacteria harboring integrons and antimicrobial resistance genes. Appl Environ Microbiol 73:6686–6690
Baquero F, Martinez JL, Canton R (2008) Antibiotics and antibiotic resistance in water environments. Curr Opin Biotechnol 19:260–265
Baquirin MH, Barlow M (2008) Evolution and recombination of the plasmidic qnr alleles. Mol Evol 67:103–110
Chen H, Shu W, Chang X, Chen JA, Guo Y, Tan Y (2010) The profile of antibiotics resistance and integrons of extended-spectrum beta-lactamase producing thermotolerant coliforms isolated from the Yangtze River basin in Chongqing. Environ Pollut 158(7):2459–2464
Cheung TK, Chu YW, Chu MY, Ma CH, Yung RW, Kam KM (2005) Plasmid-mediated resistance to ciprofloxacin and cefotaxime in clinical isolates of Salmonella enterica serotype Enteritidis in Hong Kong. Antimicrob Chemother 56:586–589
Chopra I, Roberts M (2001) Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev 65:232–260
Clinical and Laboratory Standards Institute (2004) Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically (7th ed). CLSI, Wayne
Costa D, Poeta P, Sáenz Y, Coelho AC, Matos M, Vinué L et al (2008) Prevalence of antimicrobial resistance and resistance genes in faecal Escherichia coli isolates recovered from healthy pets. Vet Microbiol 127(1–2):97–105
Farzaneh S, Peduzzi J, Sofer L, Reynaud A, Barthelemy M, Labia R (1997) Characterization and amino acid sequence of the OXY-2 group for β-lactamase of pI 57 isolated from aztreonam-resistant Klebsiella oxytoca strain HB60. J Antimicrob Chemother 40:789–795
Henriques IS, Fonseca F, Alves A, Saavedra MJ, Correia A (2006) Occurrence and diversity of integrons and β-lactamase genes among ampicillin-resistant isolates from estuarine waters. Res Microbiol 157:938–947
Hu JY, Shi JC, Chang H, Li D, Yang M, Kamagata Y (2008) Phenotyping and genotyping of antibiotic-resistant Escherichia coli isolated from a natural river basin. Environ Sci Technol 42:3415–3420
Ibekwe AM, Murinda SE, Graves AK (2011) Genetic diversity and antimicrobial resistance of Escherichia coli from human and animal sources uncovers multiple resistances from human sources. PLoS One 6:e20819
Kemper N (2008) Veterinary antibiotics in the aquatic terrestrial environment. Ecol Indic 8:1–13
Lamshöft M, Sukul P, Zühlke S, Spiteller M (2007) Metabolism of 14C-labelled and non-labelled sulfadiazine after administration to pigs. Anal Bioanal Chem 388:1733–1745
Lu SY, Zhang YL, Geng SN, Li TY, Ye ZM, Zhang DS et al (2010) High diversity of extended-spectrum beta-lactamase-producing bacteria in an urban river sediment habitat. Appl Environ Microbiol 76(17):5972–5976
Ma J, Zeng Z, Chen Z, Xu X, Wang X, Deng Y et al (2009) High prevalence of plasmid-mediated quinolone resistance determinants qnr, aac(6′)-Ib-cr, and qepA among ceftiofur-resistant Enterobacteriaceae isolates from companion and food-producing animals. Antimicrob Agents Chemother 53(2):519–524
Nordmann P, Poirel L (2005) Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. Antimicrob Chemother 56:463–469
Peak N, Knapp CW, Yang RK, Hanfelt MM, Smith MS, Aga DS et al (2007) Abundance of six tetracycline resistance genes in wastewater lagoons at cattle feedlots with different antibiotic use strategies. Environ Microbiol 9(1):143–151
Perreten V, Boerlin P (2003) A new sulfonamide resistance gene (sul3) in Escherichia coli is widespread in the pig population of Switzerland. Antimicrob Agents Chemother 47:1169–1172
Pezzella C, Ricci A, DiGiannatale E, Luzzi Ida, Carattoli A (2004) Tetracycline and streptomycin resistance genes, transposons, and plasmids in Salmonella enterica isolates from animals in Italy. Antimicrob Agents Chemother 48(3):903–908
Pitout JDD, Laupland KB (2008) Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis 8:159–166
Poirel L, Rodriguez-Martinez JM, Mammeri H, Liard A, Nordmann P (2005) Origin of plasmid-mediated resistance determinant qnrA. Antimicrob Agents Chemother 49:3523–3525
Radstrom P, Swedberg G, Skold O (1991) Genetic analyses of sulfonamide resistance and its dissemination in Gram-negative bacteria illustrate new aspects of R plasmid evolution. Antimicrob Agents Chemother 35:1840–1848
Ram S, Vajpayee P, Shanker R (2007) Prevalence of multi-antimicrobial-agent resistant, shiga toxin and enterotoxin producing Escherichia coli in surface waters of river Ganga. Environ Sci Technol 41:7383–7388
Roberts MC (1994) Epidemiology of tetracycline-resistance determinants. Trends Microbiol 2:353–357
Roberts MC (2005) Update on acquired tetracycline resistance genes. FEMS Microbiol Lett 245:195–203
Robicsek A, Sahm DF, Strahilevitz J, Jacoby GA (2005) Broader distribution of plasmid-mediated quinolone resistance in the United States. Antimicrob Agents Chemother 49:3001–3003
Robicsek A, Jacoby GA, Hooper DC (2006a) The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis 6(10):629–640
Robicsek A, Strahilevitz J, Sahm DF, Jacoby GA, Hooperm DC (2006b) qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrobial Agents Chemotherapy 50:2872–2874
Sharma J, Sharma M, Ray P (2010) Detection of TEM & SHV genes in Escherichia coli and Klebsiella pneumoniae isolates in a tertiary care hospital from India. Indian J Med Res 132:332–336
Soufi L, Abbassi MS, Sáenz Y, Vinué L, Somalo S, Zarazaga M (2009) Prevalence and diversity of integrons and associated resistance genes in Escherichia coli isolates from poultry meat in Tunisia. Foodborne Pathogen Dis 6:1067–1073
Srinivasan V, Nam HM, Sawant AA, Headrick SI, Nguyen LT, Oliver SP (2008) Distribution of tetracycline and streptomycin resistance genes and class 1 integrons in Enterobacteriaceae isolated from dairy and nondairy farm soils. Microb Ecol 55(2):184–193
Su HC, Ying GG, Tao R, Zhang RQ, Zhao JL, Liu YS (2012) Class 1 and 2 integrons, sul resistance genes and antibiotic resistance in Escherichia coli isolated from Dongjiang River, South China. Environ Pollut 169:42–49
Sunde M, Norström M (2005) The genetic background for streptomycin resistance in Escherichia coli influences the distribution of MICs. J Antimicrob Chemother 56(1):87–90
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thevenon F, Adatte T, Wildi W, Poté J (2012) Antibiotic resistant bacteria/genes dissemination in lacustrine sediments highly increased following cultural eutrophication of Lake Geneva (Switzerland). Chemosphere 86(5):468–476
Tuckman M, Petersen PJ, Howe AY, Orlowski M, Mullen S, Chan K et al (2007) Occurrence of tetracycline resistance genes among Escherichia coli isolates from the phase 3 clinical trials for tigecycline. Antimicrob Agents Chemother 51(9):3205–3211
Wilkerson C, Samadpour M, van Kirk N, Roberts MC (2004) Antibiotic resistance and distribution of tetracycline resistance genes in Escherichia coli O157:H7 isolates from humans and bovines. Antimicrob Agents Chemother 48:1066–1067
Yamane K, Wachino J, Suzuki S, Kimura K, Shibata N, Kato H et al (2007) New plasmid-mediated fluoroquinolone efflux pump, qepA, found in an Escherichia coli clinical isolate. Antimicrob Agents Chemother 51:3354–3360
Yang S, Carlson K (2003) Evolution of antibiotics occurrence in a river through pristine, urban and agricultural landscapes. Water Res 37:4645–4656
Zhang XX, Zhang T, Fang Herbert HP (2009) Antibiotic resistance genes in water environment. Appl Microbiol Biotechnol 82:397–414
Acknowledgments
We are grateful for grants from the National Program (973 Program) on Key Basic Research Project (Grant No. 2010CB429006), the National Natural Science Foundation (Key Program) of China (Grant No. 50830304), the Nonprofit Industry Financial Program of the Ministry of Water Resources (Grant No. 201101020), the Natural Science Foundation of Jiangsu Province (Grant No. BK2012413), and the Fundamental Research Funds for the Central University (Grant No. 2012B06314).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, C., Gu, X., Zhang, S. et al. Characterization of Antibiotic-Resistance Genes in Antibiotic Resistance Escherichia coli Isolates From a Lake. Arch Environ Contam Toxicol 65, 635–641 (2013). https://doi.org/10.1007/s00244-013-9932-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00244-013-9932-2