Abstract
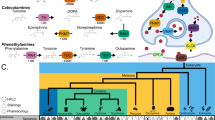
The evolution of enzyme genes at the pathway level has attracted increasing attention in recent years. Most investigations have focused on microorganisms, plants and invertebrates but rarely on vertebrates. The dopamine pathway, which participates in almost every aspect of brain function, is an excellent candidate for study at the pathway level. Herein, we report data on the divergence of six dopamine metabolic enzyme genes (three anabolic, three catabolic enzymes) and five dopamine receptor genes across five mammals, namely Homo sapiens, Pan troglodytes, Macaca mulatta, Mus musculus, and Rattus norvegicus. For enzyme genes, our data confirm previous conclusion that the upstream genes have evolved more slowly than downstream genes. Moreover, we found that catabolic genes in the dopamine metabolic pathway have evolved faster than anabolic genes, and maximum likelihood analysis suggested that this difference in evolutionary rate may be explained by anabolic genes being more constrained during selection. For dopamine receptor genes, however, the broadly expressed genes have tended to evolve more slowly than the narrowly expressed genes; maximum likelihood analysis showed that the relatively rapid evolutionary rate of the narrowly expressed receptor genes was a consequence of relaxed selective constraints. Finally, our data imply that selective constraints on synonymous sites in enzyme genes are relaxed compared with those of receptor genes because of differences in their patterns of functional regulation.


Similar content being viewed by others
References
Bairoch A et al (2009) The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res 37:D169–D174
Campillos M et al (2006) Identification and analysis of evolutionarily cohesive functional modules in protein networks. Genome Res 16:374–382
Chen YW, Dokholyan NV (2006) The coordinated evolution of yeast proteins is constrained by functional modularity. Trends Genet 22:416–419
Cork JM, Purugganan MD (2004) The evolution of molecular genetic pathways and networks. Bioessays 26:479–484
Drummond DA et al (2005) Why highly expressed proteins evolve slowly. Proc Natl Acad Sci USA 102:14338–14343
Duret L, Mouchiroud D (2000) Determinants of substitution rates in mammalian genes: expression pattern affects selection intensity but not mutation rate. Mol Biol Evol 17:68–74
Eanes WF et al (2006) Flux control and excess capacity in the enzymes of glycolysis and their relationship to flight metabolism in Drosophila melanogaster. Proc Natl Acad Sci USA 103:19413–19418
Fisher RA (1921) On the probable error of a coefficient of correlation deduced from a small sample. Metron 1:3–32
Flipphi M et al (2009) Biodiversity and evolution of primary carbon metabolism in Aspergillus nidulans and other Aspergillus spp. Fungal Genet Biol 46:S19–S44
Flowers JM et al (2007) Adaptive evolution of metabolic pathways in Drosophila. Mol Biol Evol 24:1347–1354
Fraser HB et al (2002) Evolutionary rate in the protein interaction network. Science 296:750–752
Freeman D et al (1991) Statistics. W. W. Norton & Co, New York
Gibbs RA et al (2004) Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428:493–521
Gibbs RA et al (2007) Evolutionary and biomedical insights from the rhesus macaque genome. Science 316:222–234
Hahn MW et al (2004) Molecular evolution in large genetic networks: does connectivity equal constraint? J Mol Evol 58:203–211
Hershberg R, Petrov DA (2008) Selection on codon bias. Annu Rev Genet 42:287–299
Ikemura T (1985) Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol 2:13–34
Kanehisa M et al (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36:D480–D484
Kimura M (1991) Recent development of the neutral theory viewed from the Wrightian tradition of theoretical population genetics. Proc Natl Acad Sci USA 88:5969–5973
Kitami T, Nadeau JH (2002) Biochemical networking contributes more to genetic buffering in human and mouse metabolic pathways than does gene duplication. Nat Genet 32:191–194
Larkin MA et al (2007) Clustal W and clustal X version 2.0. Bioinformatics 23:2947–2948
Levitt M, et al (1965) Elucidation of the rate-limiting step in norepinepherine biosynthesis in the perfused guinea-pig heart. J Pharmacol Exp Ther 148:1–8
Lu YQ, Rausher MD (2003) Evolutionary rate variation in anthocyanin pathway genes. Mol Biol Evol 20:1844–1853
Ma HW, et al (2007) The Edinburgh human metabolic network reconstruction and its functional analysis. Mol Syst Biol 3:135
Ma XH, Wang ZW (2009) Anticancer drug discovery in the future: an evolutionary perspective. Drug Discov Today 14:1136–1142
Mei CD et al (2009) Getting specialized: presynaptic and postsynaptic dopamine D2 receptors. Curr Opin Pharmacol 9:53–58
Mikkelsen TS et al (2005) Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437:69–87
Montague PR et al (2004) Computational roles for dopamine in behavioural control. Nature 431:760–767
Moriyama EN, Powell JR (1997) Synonymous substitution rates in Drosophila: mitochondrial versus nuclear genes. J Mol Evol 45:378–391
Pal C et al (2001) Highly expressed genes in yeast evolve slowly. Genetics 158:927–931
Park SG, Choi SS (2010) Expression breadth and expression abundance behave differently in correlations with evolutionary rates. BMC Evol Biol 10. doi:10.1186/1471-2148-1110-1241
Pereira-Leal JB et al (2006) The origins and evolution of functional modules: lessons from protein complexes. Philos Trans R Soc B Biol Sci 361:507–517
Plotkin JB et al (2004) Tissue-specific codon usage and the expression of human genes. Proc Natl Acad Sci USA 101:12588–12591
Ramsay H et al (2009) The correlation of evolutionary rate with pathway position in plant terpenoid biosynthesis. Mol Biol Evol 26:1045–1053
Rausher MD et al (1999) Patterns of evolutionary rate variation among genes of the anthocyanin biosynthetic pathway. Mol Biol Evol 16:266–274
Rausher MD et al (2008) Variation in constraint versus positive selection as an explanation for evolutionary rate variation among anthocyanin genes. J Mol Evol 67:137–144
Rozas J et al (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497
Rutter MT, Zufall RA (2004) Pathway length and evolutionary constraint in amino acid biosynthesis. J Mol Evol 58:218–224
Saeed R, Deane CM (2006) Protein–protein interactions, evolutionary rate, abundance and age. BMC Bioinformatics 7:128
Sharp PM, Li W-H (1986) Codon usage in regulatory genes in Escherichia coli does not reflect selection for ‘rare’ codons. Nucleic Acids Res 14:7737–7749
Sharp PM et al (1995) DNA sequence evolution: the sounds of silence. Philos Trans Biol Sci 349:241–247
Sims GE et al (2009) Whole-genome phylogeny of mammals: evolutionary information in genic and nongenic regions. Proc Natl Acad Sci USA 106:17077–17082
Snel B, Huynen MA (2004) Quantifying modularity in the evolution of biomolecular systems. Genome Res 14:391–397
Tamura K et al (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Velasco AM et al (2002) Molecular evolution of the lysine biosynthetic pathways. J Mol Evol 55:445–459
Vitkup D, et al (2006) Influence of metabolic network structure and function on enzyme evolution. Genome Biol 7:R39
Waterston RH et al (2002) Initial sequencing and comparative analysis of the mouse genome. Nature 420:520–562
Wong WSW et al (2004) Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics 168:1041–1051
Wright KM, Rausher MD (2010) The evolution of control and distribution of adaptive mutations in a metabolic pathway. Genetics 184:483–502
Yang ZH (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591
Yang YH, et al (2009) Evolutionary rate patterns of the Gibberellin pathway. BMC Evol Biol 9:206
Zhang A et al (2007) Recent progress in development of dopamine receptor subtype-selective agents: potential therapeutics for neurological and psychiatric disorders. Chem Rev 107:274–302
Zhao J, et al (2007) Modular co-evolution of metabolic networks. BMC Bioinformatics 8:311–322
Zuniga M et al (2002) Evolution of arginine deiminase (ADI) pathway genes. Mol Phylogenet Evol 25:429–444
Acknowledgments
We would like to thank the reviewers for their very constructive comments that help improve the manuscript. This study was supported by the National Natural Science Foundation of China (NSFC-20536040, NSFC-20875068), the National Project of Key Fundamental Research (2007CB707802), the Development Project of Science and Technology of Tianjin (05YFGZGX04500) and Programme of Introducing Talents of Discipline to Universities (B06006).
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ma, X., Wang, Z. & Zhang, X. Evolution of Dopamine-Related Systems: Biosynthesis, Degradation and Receptors. J Mol Evol 71, 374–384 (2010). https://doi.org/10.1007/s00239-010-9392-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-010-9392-5