Abstract
Diet in fish is influenced by multiple factors including nutritional requirements, trophic morphology and spatial and temporal variation in resource availability. We examined spatial variation in trophic resources on substrata grazed by scarinine parrotfishes by combining quantitative microhistology with 16S and 18S small subunit rRNA barcoding of feeding substrata in six parrotfish species on outer-shelf reefs of the Great Barrier Reef, Australia. We then compared four of these taxa with conspecific data from mid-shelf reefs differing in incident wave energy, parrotfish assemblage structure and benthic cover of hard corals, crustose coralline algae (CCA) and macroalgae. The dominant biota on outer-shelf feeding substrata in terms of both surface coverage and frequency of occurrence were filamentous cyanobacteria. The density of filamentous cyanobacteria on outer-shelf feeding substrata as measured by microscope did not differ either among the six parrotfish species or within-species cross-shelf. Endolithic and epilithic filamentous cyanobacteria from the order Nostocales were the most frequently observed filamentous cyanobacteria, suggesting that these represent a key feeding target for these parrotfishes. In addition to filamentous Nostocales cyanobacteria, taxa that were consistently present on both mid-shelf and outer-shelf feeding substrata were the euendolithic micro-chlorophytes Ostreobium and Phaeophila, diatoms, fungi, CCA, Peyssonnelia, dinoflagellates of the family Symbiodiniaceae, the sponge taxa Clionaida and Poecilosclerida and the filamentous algae Sphacelaria and Polysiphonia. Our results reveal key nutritional drivers underlying feeding by parrotfish on carbonate reefs and provide further support for the hypothesis that microscopic photoautotrophs are a major dietary target for grazing parrotfishes.









Similar content being viewed by others
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary material. Raw data are available from the corresponding author, upon reasonable request.
Change history
15 September 2023
A Correction to this paper has been published: https://doi.org/10.1007/s00227-023-04297-y
References
Ahlgren G, Gustafsson I, Boberg M (1992) Fatty acid content and chemical composition of freshwater microalgae 1. J Phycol 28:37–50
Australian Institute of Marine Science (AIMS) (2023) AIMS Long-term Monitoring Program: Benthic Community Cover Data (Great Barrier Reef).
Bellwood DR (1994) A phylogenetic study of the parrotfishes family scaridae (Pisces : Labroidei ), with a revision of genera. Rec Aust Mus 20:1–86
Bellwood DR (1995a) Carbonate transport and within-reef patterns of bioerosion and sediment release by parrotfishes (family Scaridae) on the Great Barrier Reef Carbonate transport an bioerosion and sediment release by parrotfishes (family Scaridae) on the Great Barrier Reef. Mar Ecol Prog Ser 117:127–136
Bellwood DR (1995b) Direct estimate of bioerosion by two parrotfish species, Chlorurus gibbus and C. sordidus, on the Great Barrier Reef. Australia Mar Biol 121:419–429. https://doi.org/10.1007/BF00349451
Bellwood DR (1996) Production and reworking of sediment by parrotfishes (family Scaridae) on the Great Barrier Reef, Australia. Mar Biol 125:795–800
Bellwood DR, Choat JH (1990) A functional analysis of grazing in parrotfishes (family Scaridae): the ecological implications. Environ Biol Fishes 28:189–214. https://doi.org/10.1007/BF00751035
Bellwood DR, Wainwright PC (2001) Locomotion in labrid fishes: implications for habitat use and cross-shelf biogeography on the Great Barrier Reef. Coral Reefs 20:139–150. https://doi.org/10.1007/s003380100156
Bellwood DR, Hoey AS, Choat JH (2003) Limited functional redundancy in high diversity systems: resilience and ecosystem function on coral reefs. Ecol Lett 6:281–285. https://doi.org/10.1046/j.1461-0248.2003.00432.x
Bellwood DR, Hughes TP, Folke C, Nyström M (2004) Confronting the coral reef crisis. Nature 429:827–833. https://doi.org/10.1038/nature02691
Bellwood DR, Hoey AS, Hughes TP (2012) Human activity selectively impacts the ecosystem roles of parrotfishes on coral reefs. Proc Royal Soc B Biol Sci 279:1621–1629. https://doi.org/10.1098/rspb.2011.1906
Bellwood DR, Streit RP, Brandl SJ, Tebbett SB (2019) The meaning of the term ‘function’ in ecology: a coral reef perspective. Funct Ecol 33:948–961. https://doi.org/10.1111/1365-2435.13265
Bjordal MV, Jensen KH, Sjøtun K (2020) A field study of the edible red alga Vertebrata lanosa (Rhodophyta). J Appl Phycol 32:671–681. https://doi.org/10.1007/s10811-019-01934-2
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang K Bin, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu Y-X, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik A V., Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. https://doi.org/10.1038/s41587-019-0209-9
Bonaldo RM, Bellwood DR (2011) Parrotfish predation on massive porites on the Great Barrier Reef. Coral Reefs 30:259–269. https://doi.org/10.1007/s00338-010-0669-3
Bonaldo RM, Welsh JQ, Bellwood DR (2012) Spatial and temporal variation in coral predation by parrotfishes on the GBR: evidence from an inshore reef. Coral Reefs 31:263–272. https://doi.org/10.1007/s00338-011-0846-z
Bonaldo RM, Hoey AS, Bellwood DR (2014) The ecosystem roles of parrotfishes on tropical reefs. Oceanogr Mar Biol Annu Rev 52:81–132
Brocke HJ, Polerecky L, de Beer D, Weber M, Claudet J, Nugues MM (2015) Organic matter degradation drives benthic cyanobacterial mat abundance on Caribbean Coral Reefs. PLoS ONE 10:e0125445. https://doi.org/10.1371/journal.pone.0125445
Bruggemann JH, Van Oppen MJH, Breeman AM (1994) Foraging by the stoplight parrotfish Sparisoma viride. I. Food selection in different, socially determined habitats. Mar Ecol Prog Ser 106:41–55. https://doi.org/10.3354/meps106041
Bryan PG (1975) Food habits, functional digestive morphology, and assimilation efficiency of the rabbitfish Siganus spinus (Pisces, Siganidae) on Guam. Pac Sci 29:269–277
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP (2016) DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. https://doi.org/10.1038/nmeth.3869
Callahan BJ, McMurdie PJ, Holmes SP (2017) Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J 11:2639–2643. https://doi.org/10.1038/ismej.2017.119
Casey JM, Meyer CP, Morat F, Brandl SJ, Planes S, Parravicini V (2019) Reconstructing hyperdiverse food webs: gut content metabarcoding as a tool to disentangle trophic interactions on coral reefs. Methods Ecol Evol 10:1157–1170. https://doi.org/10.1111/2041-210X.13206
Castro-Sanguino C, Sánchez JA (2012) Dispersal of Symbiodinium by the stoplight parrotfish Sparisoma viride. Biol Lett 8:282–286. https://doi.org/10.1098/rsbl.2011.0836
Charendoff JA, Edwards CB, Pedersen NE, Petrovic V, Zgliczynski B, Sandin SA, Smith JE (2023) Variability in composition of parrotfish bite scars across space and over time on a central Pacific atoll. Coral Reefs. https://doi.org/10.1007/s00338-023-02392-6
Chartock MA (1983) The role of Acanthurus guttatus (Bloch and Schneider 1801) in cycling algal production to detritus. Biotropica 15:117–121
Cheal A, Emslie M, Miller I, Sweatman H (2012) The distribution of herbivorous fishes on the Great Barrier Reef. Mar Biol 159:1143–1154. https://doi.org/10.1007/s00227-012-1893-x
Chen YC (2012) The biomass and total lipid content and composition of twelve species of marine diatoms cultured under various environments. Food Chem 131:211–219. https://doi.org/10.1016/j.foodchem.2011.08.062
Choat JH, Clements KD, Robbins W (2002) The trophic status of herbivorous fishes on coral reefs. Mar Biol 140:613–623. https://doi.org/10.1007/s00227-001-0715-3
Clare EL (2014) Molecular detection of trophic interactions: emerging trends, distinct advantages, significant considerations and conservation applications. Evol Appl 7:1144–1157. https://doi.org/10.1111/eva.12225
Clements KD, Bellwood DR (1988) A comparison of the feeding mechanisms of two herbivorous labroid fishes, the temperate Odax pullus and the tropical Scarus rubroviolaceus. Mar Freshw Res 39:87. https://doi.org/10.1071/MF9880087
Clements KD, Choat JH (2018) Nutritional ecology of parrotfishes (Scarinae, Labridae). In: Hoey AS, Bonaldo RM (eds) Biology of parrotfishes. CRC Press, Boca Raton, FL, pp 42–68
Clements KD, German DP, Piché J, Tribollet A, Choat JH (2017) Integrating ecological roles and trophic diversification on coral reefs: multiple lines of evidence identify parrotfishes as microphages. Biol J Lin Soc 120:729–751. https://doi.org/10.1111/bij.12914
Coker DJ, DiBattista JD, Stat M, Arrigoni R, Reimer J, Terraneo TI, Villalobos R, Nowicki JP, Bunce M, Berumen ML (2022) DNA metabarcoding confirms primary targets and breadth of diet for coral reef butterflyfishes. Coral Reefs 42:1–15. https://doi.org/10.1007/s00338-022-02302-2
Crossman D, Choat J, Clements K (2005) Nutritional ecology of nominally herbivorous fishes on coral reefs. Mar Ecol Prog Ser 296:129–142. https://doi.org/10.3354/meps296129
Curren E, Leaw CP, Lim PT, Leong SCY (2022) The toxic cosmopolitan cyanobacteria Moorena producens: insights into distribution, ecophysiology and toxicity. Environ Sci Pollut Res 29:78178–78206. https://doi.org/10.1007/s11356-022-23096-4
Dalsgaard J, St. John M, Kattner G, Müller-Navarra D, Hagen W (2003) Fatty acid trophic markers in the pelagic marine environment. In: Advances in marine biology, pp 225–340
Dawson EY, Aleem AA, Halstead BW (1955) Marine algae from Palmyra Island with special reference to the feeding habits and toxicology of reef fishes. University of Southern California Press, Los Angeles
Deagle BE, Thomas AC, McInnes JC, Clarke LJ, Vesterinen EJ, Clare EL, Kartzinel TR, Eveson JP (2019) Counting with DNA in metabarcoding studies: how should we convert sequence reads to dietary data? Mol Ecol 28:391–406. https://doi.org/10.1111/mec.14734
Dean AJ, Steneck RS, Tager D, Pandolfi JM (2015) Distribution, abundance and diversity of crustose coralline algae on the Great Barrier Reef. Coral Reefs 34:581–594. https://doi.org/10.1007/s00338-015-1263-5
Deinhart M, Mills MS, Schils T (2022) Community assessment of crustose calcifying red algae as coral recruitment substrates. PLoS ONE 17:e0271438. https://doi.org/10.1371/journal.pone.0271438
Desbiens A, Roff G, Razak TB, Wolfe K, Emslie MJ, Hoey A, Kennedy E V, Thompson A, Adams MP, Baird M, Chaloupka M, Diaz-Pulido G, Mongin M, Mumby PJ (2021) Integrative carbonate budget model of the Great Barrier Reef Report to the National Environmental Science Program. Reef and Rainforest RFesearch Centre Limited, Cairns (23pp.).
Dunlap M, Pawlik JR (1998) Spongivory by Parrotfish in Florida Mangrove and Reef Habitats. Mar Ecol 19:325–337. https://doi.org/10.1111/j.1439-0485.1998.tb00471.x
Dunstan GA, Volkman JK, Barrett SM, Leroi J-M, Jeffrey SW (1993) Essential polyunsaturated fatty acids from 14 species of diatom (Bacillariophyceae). Phytochemistry 35:155–161. https://doi.org/10.1016/S0031-9422(00)90525-9
Edmunds PJ, Zimmermann SA, Bramanti L (2019) A spatially aggressive peyssonnelid algal crust (PAC) threatens shallow coral reefs in St. John. US Virgin Islands Coral Reefs 38:1329–1341. https://doi.org/10.1007/s00338-019-01846-0
Eggertsen M, Chacin DH, van Lier J, Eggertsen L, Fulton CJ, Wilson S, Halling C, Berkström C (2020) Seascape configuration and fine-scale habitat complexity shape parrotfish distribution and function across a Coral Reef Lagoon. Diversity 12:391. https://doi.org/10.3390/d12100391
Fabricius KE, Crossman K, Jonker M, Mongin M, Thompson A (2023) Macroalgal cover on coral reefs: spatial and environmental predictors, and decadal trends in the Great Barrier Reef. PLoS ONE 18:e0279699. https://doi.org/10.1371/journal.pone.0279699
Fey P, Parravicini V, Bănaru D, Dierking J, Galzin R, Lebreton B, Meziane T, Polunin NVC, Zubia M, Letourneur Y (2021) Multi-trophic markers illuminate the understanding of the functioning of a remote, low coral cover Marquesan coral reef food web. Sci Rep 11:20950. https://doi.org/10.1038/s41598-021-00348-w
Foster JS, Green SJ, Ahrendt SR, Golubic S, Reid RP, Hetherington KL, Bebout L (2009) Molecular and morphological characterization of cyanobacterial diversity in the stromatolites of Highborne Cay, Bahamas. ISME J 3:573–587. https://doi.org/10.1038/ismej.2008.129
Frantz CM, Petryshyn VA, Corsetti FA (2015) Grain trapping by filamentous cyanobacterial and algal mats: implications for stromatolite microfabrics through time. Geobiology 13:409–423. https://doi.org/10.1111/gbi.12145
Frydl P, Stearn CW (1978) Rate of bioerosion by parrotfish in Barbados reef environments. J Sediment Res. https://doi.org/10.1306/212F7612-2B24-11D7-8648000102C1865D
Fujise L, Suggett DJ, Stat M, Kahlke T, Bunce M, Gardner SG, Goyen S, Woodcock S, Ralph PJ, Seymour JR, Siboni N, Nitschke MR (2021) Unlocking the phylogenetic diversity, primary habitats, and abundances of free-living Symbiodiniaceae on a coral reef. Mol Ecol 30:343–360. https://doi.org/10.1111/mec.15719
Gaino E, Sarà M (1994) Siliceous spicules of Tethya seychellensis (Porifera) support the growth of a green alga: a possible light conducting system. Mar Ecol Prog Ser 108:147–152. https://www.int-res.com/articles/meps/108/m108p147
Garcia-Pichel F, López-Cortés A, Nübel U (2001) Phylogenetic and morphological diversity of cyanobacteria in soil desert crusts from the Colorado plateau. Appl Environ Microbiol 67:1902–1910. https://doi.org/10.1128/AEM.67.4.1902-1910.2001
Garnick S, Barboza PS, Walker JW (2018) Assessment of animal-based methods used for estimating and monitoring rangeland herbivore diet composition. Rangel Ecol Manag 71:449–457. https://doi.org/10.1016/j.rama.2018.03.003
Genner MJ, Turner GF, Hawkins SJ (1999) Foraging of rocky habitat cichlid fishes in Lake Malawi: coexistence through niche partitioning? Oecologia 121:283–292
Goatley CHR, Bellwood DR (2012) Sediment suppresses herbivory across a coral reef depth gradient. Biol Lett 8:1016–1018. https://doi.org/10.1098/rsbl.2012.0770
Gobalet KW (2018) Cranial specializations of parrotfishes, genus Scarus (Scarinae, Labridae) for scraping reef surfaces. In: Andrew SH, Roberta MB, Andrew SH, Roberta MB (eds) Biology of parrotfishes. CRC Press, Boca Raton, pp 1–10
Goldberg EG, Raab TK, Desalles P, Briggs AA, Dunbar RB, Millero FJ, Woosley RJ, Young HS, Micheli F, Mccauley DJ (2019) Chemistry of the consumption and excretion of the bumphead parrotfish (Bolbometopon muricatum), a coral reef mega-consumer. Coral Reefs 38:347–357. https://doi.org/10.1007/s00338-019-01781-0
Golubic S, Campbell SE (1981) Biogenically formed aragonite concretions in marine rivularia. Phanerozoic stromatolites. Springer, Berlin Heidelberg, Berlin, Heidelberg, pp 209–229
González-Murcia S, Ekins M, Bridge TCL, Battershill CN, Jones GP (2023) Substratum selection in coral reef sponges and their interactions with other benthic organisms. Coral Reefs. https://doi.org/10.1007/s00338-023-02350-2
Gordon SE, Goatley CHR, Bellwood DR (2016) Low-quality sediments deter grazing by the parrotfish Scarus rivulatus on inner-shelf reefs. Coral Reefs 35:285–291. https://doi.org/10.1007/s00338-015-1374-z
Grange JS, Rybarczyk H, Tribollet A (2015) The three steps of the carbonate biogenic dissolution process by microborers in coral reefs (New Caledonia). Environ Sci Pollut Res 22:13625–13637. https://doi.org/10.1007/s11356-014-4069-z
Grupstra CGB, Rabbitt KM, Howe-Kerr LI, Correa AMS (2021) Fish predation on corals promotes the dispersal of coral symbionts. Anim Microbiome 3:25. https://doi.org/10.1186/s42523-021-00086-4
Guiry MD, Guiry GM (2022) AlgaeBase. World-wide electronic publication. National University of Ireland, Galway
Gust N, Choat JH, Ackerman JL (2002) Demographic plasticity in tropical reef fishes. Mar Biol 140:1039–1051. https://doi.org/10.1007/s00227-001-0773-6
Harris PM, Halley RB, Lukas KJ (1979) Endolith microborings and their preservation in Holocene-Pleistocene (Bahama-Florida) ooids. Geology 7:216. https://doi.org/10.1130/0091-7613(1979)7%3c216:EMATPI%3e2.0.CO;2
Hata H, Watanabe K, Kato M (2010) Geographic variation in the damselfish-red alga cultivation mutualism in the Indo-West Pacific. BMC Evol Biol 10:185
Hata H, Takano S, Masuhara H (2020) Herbivorous damselfishes expand their territories after causing white scars on Porites corals. Sci Rep 10:16172. https://doi.org/10.1038/s41598-020-73232-8
Herlemann DP, Labrenz M, Jürgens K, Bertilsson S, Waniek JJ, Andersson AF (2011) Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J 5:1571–1579. https://doi.org/10.1038/ismej.2011.41
Hill M, Allenby A, Ramsby B, Schönberg C, Hill A (2011) Symbiodinium diversity among host clionaid sponges from Caribbean and Pacific reefs: evidence of heteroplasmy and putative host-specific symbiont lineages. Mol Phylogenet Evol 59:81–88. https://doi.org/10.1016/j.ympev.2011.01.006
Hoey AS, Bellwood DR (2008) Cross-shelf variation in the role of parrotfishes on the Great Barrier Reef. Coral Reefs 27:37–47. https://doi.org/10.1007/s00338-007-0287-x
Homma C, Inokuchi D, Nakamura Y, Ohnishi K, Funaki H, Yamaguchi H, Adachi M (2022) Comparison of the diets of the parrotfishes Scarus ovifrons and Calotomus japonicus using rDNA metabarcoding. Fish Sci 88:539–553. https://doi.org/10.1007/s12562-022-01623-z
Hopley D (2006) Coral Reef growth on the shelf margin of the Great Barrier Reef with special reference to the pompey complex. J Coast Res 221:150–158. https://doi.org/10.2112/05A-0012.1
Huertas V, Morais RA, Bonaldo RM, Bellwood DR (2021) Parrotfish corallivory on stress-tolerant corals in the Anthropocene. PLoS ONE 16:e0250725. https://doi.org/10.1371/journal.pone.0250725
Johansen JR, González‐Resendiz L, Escobar‐Sánchez V, Segal‐Kischinevzky C, Martínez‐Yerena J, Hernández‐Sánchez J, Hernández‐Pérez G, León‐Tejera H (2021) When will taxonomic saturation be achieved? A case study in Nunduva and Kyrtuthrix (Rivulariaceae, Cyanobacteria). J Phycol 57:1699–1720. https://doi.org/10.1111/jpy.13201
Johnson G, Taylor B, Robbins W, Franklin E, Toonen R, Bowen B, Choat J (2019) Diversity and structure of parrotfish assemblages across the Northern Great Barrier Reef. Diversity 11:14. https://doi.org/10.3390/d11010014
Jones RS (1968) Ecological relationships in Hawaiian and Johnston Island Acanthuridae (surgeonfishes). Micronesica 4:309–361
Jones GP, Santana L, McCook LJ, McCormick MI (2006) Resource use and impact of three herbivorous damselfishes on coral reef communities. Mar Ecol Prog Ser 328:215–224
Kartzinel TR, Chen PA, Coverdale TC, Erickson DL, Kress WJ, Kuzmina ML, Rubenstein DI, Wang W, Pringle RM (2015) DNA metabarcoding illuminates dietary niche partitioning by African large herbivores. Proc Natl Acad Sci 112:8019–8024. https://doi.org/10.1073/pnas.1503283112
Kassambara A (2020) rstatix: pipe-friendly framework for basic statistical tests.
Katoh K, Standley DM (2013) MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Kayal E, Bentlage B, Sabrina Pankey M, Ohdera AH, Medina M, Plachetzki DC, Collins AG, Ryan JF (2018) Phylogenomics provides a robust topology of the major cnidarian lineages and insights on the origins of key organismal traits. BMC Evol Biol 18:68. https://doi.org/10.1186/s12862-018-1142-0
Kench PS (1998) Physical processes in an Indian Ocean atoll. Coral Reefs 17:155–168
Kench PS, Beetham EP, Turner T, Morgan KM, Owen SD, RogerF McLean (2022) Sustained coral reef growth in the critical wave dissipation zone of a Maldivian atoll. Commun Earth Environ 3:9. https://doi.org/10.1038/s43247-021-00338-w
Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:e1. https://doi.org/10.1093/nar/gks808
Kohler KE, Gill SM (2006) Coral point count with excel extensions (CPCe): a visual basic program for the determination of coral and substrate coverage using random point count methodology. Comput Geosci 32:1259–1269. https://doi.org/10.1016/j.cageo.2005.11.009
Komárek J, Johansen JR (2015) Filamentous cyanobacteria. In: Wehr JD, Sheath RG, Kociolek JP (eds) Freshwater algae of North America, 2nd edn. Elsevier, Boston, pp 135–235
Konstantinou D, Gerovasileiou V, Voultsiadou E, Gkelis S (2018) Sponges-Cyanobacteria associations: Global diversity overview and new data from the Eastern Mediterranean. PLoS One 13:e0195001. https://doi.org/10.1371/journal.pone.0195001
Konstantinou D, Voultsiadou E, Panteris E, Gkelis S (2021) Revealing new sponge-associated cyanobacterial diversity: Novel genera and species. Mol Phylogenet Evol 155:106991. https://doi.org/10.1016/j.ympev.2020.106991
Krassowski M (2020) ComplexUpset: Create Complex UpSet Plots Using ggplot2 Components. In: R package Version 0.5
Kuffner IB, Paul VJ (2004) Effects of the benthic cyanobacterium Lyngbya majuscula on larval recruitment of the reef corals Acropora surculosa and Pocillopora damicornis. Coral Reefs 23:455–458. https://doi.org/10.1007/s00338-004-0416-8
LaJeunesse TC, Parkinson JE, Gabrielson PW, Jeong HJ, Reimer JD, Voolstra CR, Santos SR (2018) Systematic revision of symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr Biol 28:2570-2580.e6. https://doi.org/10.1016/j.cub.2018.07.008
Lange ID, Perry CT, Stuhr M (2022) Recovery trends of reef carbonate budgets at remote coral atolls 6 years post-bleaching. Limnol Oceanogr. https://doi.org/10.1002/lno.12066
Letunic I, Bork P (2021) Interactive Tree Of Life (iTOL) v6: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–W296
Lex A, Gehlenborg N, Strobelt H, Vuillemot R, Pfister H (2014) UpSet: visualization of intersecting sets. IEEE Trans vis Comput Graph 20:1983–1992. https://doi.org/10.1109/TVCG.2014.2346248
Machida RJ, Knowlton N (2012) PCR Primers for Metazoan Nuclear 18S and 28S Ribosomal DNA Sequences. PLoS One 7:e46180. https://doi.org/10.1371/journal.pone.0046180
Mallela J, Fox RJ (2018) The role of parrotfishes in the destruction and construction of coral reefs. In: Hoey AS, Bonaldo RM (eds) Biology of parrotfishes. CRC Press, Bolca Raton, FL, pp 161–196
Mallela J, Perry CT (2007) Calcium carbonate budgets for two coral reefs affected by different terrestrial runoff regimes, Rio Bueno, Jamaica. Coral Reefs 26:129–145. https://doi.org/10.1007/s00338-006-0169-7
Mapstone BD (Bruce D, Ayling T, Choat JH, Great Barrier Reef Marine Park Authority. (1998) Habitat, cross shelf and regional patterns in the distributions and abundances of some coral reef organisms on the northern Great Barrier Reef, with comment on the implications for future monitoring. Great Barrier Reef Marine Park Authority
Marlow J, Schönberg CHL, Davy SK, Haris A, Jompa J, Bell JJ (2019) Bioeroding sponge assemblages: the importance of substrate availability and sediment. J Mar Biol Assoc UK 99:343–358. https://doi.org/10.1017/S0025315418000164
McArdle B, Anderson M (2001) Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82:290–297. https://doi.org/10.1890/0012-9658(2001)082[0290:FMMTCD]2.0.CO;2
McCauley DJ, Young HS, Guevara R, Williams GJ, Power EA, Dunbar RB, Bird DW, Durham WH, Micheli F (2014) Positive and negative effects of a threatened parrotfish on reef ecosystems. Conserv Biol 28:1312–1321. https://doi.org/10.1111/cobi.12314
McClure E, Richardson L, Graba-Landry A, Loffler Z, Russ G, Hoey A (2019) Cross-shelf differences in the response of herbivorous fish assemblages to severe environmental disturbances. Diversity 11:23. https://doi.org/10.3390/d11020023
McMurdie PJ, Holmes S (2013) Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. https://doi.org/10.1371/journal.pone.0061217
Mendes TC, Ferreira CEL, Clements KD (2018) Discordance between diet analysis and dietary macronutrient content in four nominally herbivorous fishes from the Southwestern Atlantic. Mar Biol 165:180. https://doi.org/10.1007/s00227-018-3438-4
Molina-Hernández A, González-Barrios FJ, Perry CT, Álvarez-Filip L (2020) Two decades of carbonate budget change on shifted coral reef assemblages: are these reefs being locked into low net budget states? Proc Royal Soc B: Biol Sci 287:20202305. https://doi.org/10.1098/rspb.2020.2305
Momper LM, Reese BK, Carvalho G, Lee P, Webb EA (2015) A novel cohabitation between two diazotrophic cyanobacteria in the oligotrophic ocean. ISME J 9:882–893. https://doi.org/10.1038/ismej.2014.186
Montgomery WL, Gerking SD (1980) Marine macroalgae as foods for fishes: an evaluation of potential food quality. Environ Biol Fishes 5:143–153. https://doi.org/10.1007/BF02391621
Montgomery WL, Umino T, Nakagawa H, Vaughn I, Shibuno T (1999) Lipid storage and composition in tropical surgeonfishes (Teleostei: Acanthuridae). Mar Biol 133:137–144
Morgan KM, Kench PS (2016) Parrotfish erosion underpins reef growth, sand talus development and island building in the Maldives. Sediment Geol 341:50–57. https://doi.org/10.1016/j.sedgeo.2016.05.011
Mote S, Schönberg CHL, Samaai T, Gupta V, Ingole B (2019) A new clionaid sponge infests live corals on the west coast of India (Porifera, Demospongiae, Clionaida). Syst Biodivers 17:190–206. https://doi.org/10.1080/14772000.2018.1513430
Mumby PJ, Steneck RS (2018) Paradigm lost: dynamic nutrients and missing detritus on coral reefs. Bioscience 68:487–495. https://doi.org/10.1093/biosci/biy055
Mumby PJ, Dahlgren CP, Harborne AR, Kappel CV, Micheli F, Brumbaugh DR, Holmes KE, Mendes JM, Broad K, Sanchirico JN, Buch K, Box S, Stoffle RW, Gill AB (2006) Fishing, trophic cascades, and the process of grazing on coral reefs. Science 311:98–101. https://doi.org/10.1126/science.1121129
Murray RL, Mitsui A (1982) Growth of hybrid Tilapia fry fed nitrogen fixing marine blue-green algae in seawater. Journal of the World Mariculture Soc 13:198–209. https://doi.org/10.1111/j.1749-7345.1982.tb00027.x
Nakagawa H, Umino T, Sekimoto T, Ambas I, Montgomery L, Nakano T (2002) Characterization of the digestive tract of wild ayu. Fish Sci 68:341–346. https://doi.org/10.1046/j.1444-2906.2002.00431.x
Nalley EM, Donahue MJ, Heenan A, Toonen RJ (2021) Quantifying the diet diversity of herbivorous coral reef fishes using systematic review and DNA metabarcoding. Environ DNA 00:1–15. https://doi.org/10.1002/edn3.247
Nicholson GM, Clements KD (2020) Resolving resource partitioning in parrotfishes (Scarini) using microhistology of feeding substrata. Coral Reefs 39:1313–1327. https://doi.org/10.1007/s00338-020-01964-0
Nicholson GM, Clements KD (2021) Ecomorphological divergence and trophic resource partitioning in 15 syntopic Indo-Pacific parrotfishes (Labridae: Scarini). Biol J Lin Soc 132:590–611. https://doi.org/10.1093/biolinnean/blaa210
Nicholson GM, Clements KD (2022) Scarus spinus, crustose coralline algae and cyanobacteria: an example of dietary specialization in the parrotfishes. Coral Reefs 41:1465–1479. https://doi.org/10.1007/s00338-022-02295-y
Nicholson GM, Clements KD (2023) Micro-photoautotroph predation as a driver for trophic niche specialization in 12 syntopic Indo-Pacific parrotfish species. Biol J Linnean Soc. https://doi.org/10.1093/biolinnean/blad005/7115621
Nielsen JM, Clare EL, Hayden B, Brett MT, Kratina P (2017) Diet tracing in ecology: method comparison and selection. Methods Ecol Evol 9:278–291. https://doi.org/10.1111/2041-210X.12869
Nitschke MR, Davy SK, Ward S (2016) Horizontal transmission of Symbiodinium cells between adult and juvenile corals is aided by benthic sediment. Coral Reefs 35:335–344. https://doi.org/10.1007/s00338-015-1349-0
Nitschke MR, Craveiro SC, Brandão C, Fidalgo C, Serôdio J, Calado AJ, Frommlet JC (2020) Description of Freudenthalidium gen. nov. and Halluxium gen. nov. to formally recognize Clades Fr3 and H as genera in the family Symbiodiniaceae (Dinophyceae). J Phycol 56:923–940. https://doi.org/10.1111/jpy.12999
Nitschke MR, Rosset SL, Oakley CA, Gardner SG, Camp EF, Suggett DJ, Davy SK (2022) The diversity and ecology of Symbiodiniaceae: A traits-based review. In: Advances in Marine Biology. Academic Press, pp 55–127
Odum HT, Odum EP (1955) Trophic Structure and Productivity of a Windward Coral Reef Community on Eniwetok Atoll. Ecol Monogr 25:291–320
Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P (2019) vegan: Community Ecology Package. R package version 2.5–6. 2019
Olsson J, Toth GB, Albers E (2020) Biochemical composition of red, green and brown seaweeds on the Swedish west coast. J Appl Phycol 32:3305–3317. https://doi.org/10.1007/s10811-020-02145-w
Pasella MM, Lee M-FE, Marcelino VR, Willis A, Verbruggen H (2022) Ten Ostreobium (Ulvophyceae) strains from Great Barrier Reef corals as a resource for algal endolith biology and genomics. Phycologia 1–7
Paul V, Nelson S, Sanger H (1990) Feeding preferences of adult and juvenile rabbitfish Siganus argenteus in relation to chemical defenses of tropical seaweeds. Mar Ecol Prog Ser 60:23–34. https://doi.org/10.3354/meps060023
Paul VJ, Freeman CJ, Agarwal V (2019) Chemical ecology of marine sponges: new opportunities through “-Omics.” Integr Comp Biol 59:765–776. https://doi.org/10.1093/icb/icz014
Pentecost A, Whitton BA (2012) Subaerial cyanobacteria. In: Whitton BA (ed) Ecology of cyanobacteria II. Springer, pp 291–316
Pernice M, Raina J-B, Rädecker N, Cárdenas A, Pogoreutz C, Voolstra CR (2020) Down to the bone: the role of overlooked endolithic microbiomes in reef coral health. ISME J 14:325–334. https://doi.org/10.1038/s41396-019-0548-z
Perry CT, Edinger EN, Kench PS, Murphy GN, Smithers SG, Steneck RS, Mumby PJ (2012) Estimating rates of biologically driven coral reef framework production and erosion: a new census-based carbonate budget methodology and applications to the reefs of Bonaire. Coral Reefs 31:853–868. https://doi.org/10.1007/s00338-012-0901-4
Perry CT, Kench PS, O’Leary MJ, Morgan KM, Januchowski-Hartley F (2015) Linking reef ecology to island building: parrotfish identified as major producers of island-building sediment in the Maldives. Geology 43:503–506. https://doi.org/10.1130/G36623.1
Perry CT, Salter MA, Lange ID, Kochan DP, Harborne AR, Graham NAJ (2022) Geo-ecological functions provided by coral reef fishes vary among regions and impact reef carbonate cycling regimes. Ecosphere. https://doi.org/10.1002/ecs2.4288
Pessarrodona A, Tebbett SB, Bosch NE, Bellwood DR, Wernberg T (2022) High herbivory despite high sediment loads on a fringing coral reef. Coral Reefs 41:161–173. https://doi.org/10.1007/s00338-021-02211-w
Pompanon F, Deagle B, Symondson W, Brown D, Jarman S, Taberlet P (2012) Who is eating what: diet assessment using next generation sequencing. Mol Ecol 21:1931–1950. https://doi.org/10.1111/j.1365-294X.2011.05403.x
Power ME, Stewart AJ, Matthews WJ (1988) Grazer control of algae in an Ozark mountain stream: effects of short-term exclusion. Ecology 69:1894–1898. https://doi.org/10.2307/1941166
Price I, Scott F (1992) The turf algal flora of the Great Barrier Reef part I. Rhodophyta. James Cook University of North Queensland, Townsville
Price MN, Dehal PS, Arkin AP (2010) FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. https://doi.org/10.1093/nar/gks1219
R Core Development Team (2019) R: A language and environment for statistical computing.
Radtke G, Golubic S (2011) Microbial euendolithic assemblages and microborings in intertidal and shallow marine habitats: insight in cyanobacterial speciation. In: Reitner J, Quéric N-V, Arp G (eds) Advances in stromatolite geobiology. Springer, Berlin, pp 233–263
Randall JE (1955) Fishes of the Gilbert Islands. Atoll Res Bull 47:1–243. https://doi.org/10.5479/si.00775630.47.1
Reinthal PN (1990) The feeding habits of a group of herbivorous rock-dwelling cichlid fishes (Cichlidae: Perciformes) from Lake Malawi, Africa. Environ Biol Fishes 27:215–233. https://doi.org/10.1007/BF00001674
Roush D, Giraldo-Silva A, Garcia-Pichel F (2021) Cydrasil 3, a curated 16S rRNA gene reference package and web app for cyanobacterial phylogenetic placement. Sci Data 8:1–6. https://doi.org/10.1038/s41597-021-01015-5
Rützler K (1990) Associations between Caribbean sponges and photosynthetic organisms. In: Rützler K (ed) New perspectives in sponge biology. Smithsonian Institution Press, Washington, D.C., p 533
Scoffin TP, Stearn CW, Boucher D, Frydl P, Hawkins CM, Hunter IG, Macgeachy JK (1980) Calcuim-carbonate budget of a fringing reef on the west coast of Barbados. Part 2: erosion, sediments and internal structure. Bull Mar Sci 30:475–508
Sihvonen LM, Lyra C, Fewer DP, Rajaniemi-Wacklin P, Lehtimäki JM, Wahlsten M, Sivonen K (2007) Strains of the cyanobacterial genera Calothrix and Rivularia isolated from the Baltic sea display cryptic diversity and are distantly related to Gloeotrichia and Tolypothrix. FEMS Microbiol Ecol 61:74–84
Siqueira AC, Bellwood DR, Cowman PF (2019) The evolution of traits and functions in herbivorous coral reef fishes through space and time. Proc Royal Soc b Biol Sci 286:20182672. https://doi.org/10.1098/rspb.2018.2672
Stal LJ (2012) Cyanobacterial mats and stromatolites. In: Whitton B (ed) Ecology of cyanobacteria II. Springer, Dordrecht, pp 65–125
Steneck RS (1983) Escalating herbivory and resulting adaptive trends in calcareous algal crusts. Paleobiology 9:44–61. https://doi.org/10.1017/S0094837300007375
Steneck RS (1985) Adaptations of crustose coralline algae to herbivory: patterns in space and time. Paleoalgology. https://doi.org/10.1007/978-3-642-70355-3_29
Steneck RS (1986) The ecology of coralline algal crusts: convergent patterns and adaptative strategies. Annu Rev Ecol Syst 17:273–303. https://doi.org/10.1146/annurev.es.17.110186.001421
Streelman JT, Alfaro M, Westneat MW, Bellwood DR, Karl SA (2002) Evolutionary history of the parrotfishes: biogeography, ecomorphology, and comparative diversity. Evolution 56:961–971. https://doi.org/10.1554/0014-3820(2002)056
Tebbett SB, Goatley CHR, Bellwood DR (2017a) Algal Turf sediments and sediment production by parrotfishes across the continental shelf of the Northern Great Barrier Reef. PLoS ONE 12:e0170854. https://doi.org/10.1371/journal.pone.0170854
Tebbett SB, Goatley CHR, Bellwood DR (2017b) Fine sediments suppress detritivory on coral reefs. Mar Pollut Bull. https://doi.org/10.1016/j.marpolbul.2016.11.016
Tillett D, Neilan BA (2000) Xanthogenate nucleic acid isolation from cultured and environmental cyanobacteria. J Phycol 36:251–258. https://doi.org/10.1046/j.1529-8817.2000.99079.x
Titlyanov AE, Titlyanova VT, Li X, Huang H (2017) Coral reef marine plants of Hainan island. Academic Press, London
Tribollet A (2008) Dissolution of dead corals by euendolithic microorganisms across the Northern Great Barrier Reef (Australia). Microb Ecol 55:569–580. https://doi.org/10.1007/s00248-007-9302-6
Tribollet A, Golubic S (2005) Cross-shelf differences in the pattern and pace of bioerosion of experimental carbonate substrates exposed for 3 years on the northern Great Barrier Reef, Australia. Coral Reefs 24:422–434. https://doi.org/10.1007/s00338-005-0003-7
Tribollet A, Payri C (2001) Bioerosion of the coralline alga Hydrolithon onkodes by microborers in the coral reefs of Moorea, French Polynesia. Oceanol Acta 24:329–342. https://doi.org/10.1016/S0399-1784(01)01150-1
Tribollet A, Langdon C, Golubic S, Atkinson M (2006) Endolithic microflora are major primary producers in dead carbonate substrates of Hawaiian coral reefs. J Phycol 42:292–303. https://doi.org/10.1111/j.1529-8817.2006.00198.x
Tsuda RT, Larson HK, Lu RJ (1972) Algal growth on beaks of live parrotfishes. Pac Sci 26:20–23
Urrejola C, Alcorta J, Salas L, Vásquez M, Polz MF, Vicuña R, Díez B (2019) Genomic features for desiccation tolerance and sugar biosynthesis in the extremophile Gloeocapsopsis sp. UTEX B3054. Front Microbiol 10:950. https://doi.org/10.3389/fmicb.2019.00950
Valentini A, Miquel C, Nawaz MA, Bellemain E, Coissac E, Pompanon F, Gielly L, Cruaud C, Nascetti G, Wincker P, Swenson JE, Taberlet P (2009) New perspectives in diet analysis based on DNA barcoding and parallel pyrosequencing: the trn L approach. Mol Ecol Res 9:51–60. https://doi.org/10.1111/j.1755-0998.2008.02352.x
van der Windt N, van der Ent E, Ambo-Rappe R, de Voogd NJ (2020) Presence and genetic identity of symbiodiniaceae in the bioeroding sponge Genera Cliona and Spheciospongia (Clionaidae) in the Spermonde Archipelago (SW Sulawesi), Indonesia. Front Ecol Evol. https://doi.org/10.3389/fevo.2020.595452
Veron JEN, Hudson RCL (1978) Ribbon reefs of the northern region. Philos Transac Royal Soc London b Biol Sci 284:3–21
Wainwright PC, Bellwood DR, Westneat MW, Grubich JR, Hoey AS (2004) A functional morphospace for the skull of labrid fishes: patterns of diversity in a complex biomechanical system. Biol J Lin Soc 82:1–25. https://doi.org/10.1111/j.1095-8312.2004.00313.x
Wiebe WJ, Johannes RE, Webb KL (1975) Nitrogen fixation in a coral reef community. Science. https://doi.org/10.1126/science.188.4185.257
Willenz P, Hartman WD (1989) Micromorphology and ultrastructure of Caribbean sclerosponges. Mar Biol 103:387–401. https://doi.org/10.1007/BF00397274
Wilson SK, Bellwood DR, Howard Choat J, Furnas MJ, Wilson SK, Bellwood DR, Choat JH, Furnas MJ (2003) Detritus in the epilithic algal matrix and its use by coral reef fishes. Oceanogr Mar Biol Annu Rev 41:279–309
Wolfe K, Anthony K, Babcock RC, Bay L, Bourne DG, Burrows D, Byrne M, Deaker DJ, Diaz-Pulido G, Frade PR, Gonzalez-Rivero M, Hoey A, Hoogenboom M, McCormick M, Ortiz J-C, Razak T, Richardson AJ, Roff G, Sheppard-Brennand H, Stella J, Thompson A, Watson S-A, Webster N, Audas D, Beeden R, Carver J, Cowlishaw M, Dyer M, Groves P, Horne D, Thiault L, Vains J, Wachenfeld D, Weekers D, Williams G, Mumby PJ (2020) Priority species to support the functional integrity of coral reefs. In: Hawkins SJ, Allcock AL, Bates AE, Evans AJ, Firth LB, McQuaid CD, Russell BD, Smith IP, Swearer SE, Todd PA (eds) Oceanography and marine biology. CRC Press, Boca Raton, pp 179–326
Wulff JL (2021) Targeted predator defenses of sponges shape community organization and tropical marine ecosystem function. Ecol Monogr 91:1–29. https://doi.org/10.1002/ecm.1438
Yarlett RT, Perry CT, Wilson RW (2021) Quantifying production rates and size fractions of parrotfish-derived sediment: a key functional role on Maldivian coral reefs. Ecol Evol 11:16250–16265. https://doi.org/10.1002/ece3.8306
Zubia M, Vieira C, Palinska KA, Roué M, Gaertner J-C, Zloch I, Grellier M, Golubic S (2019) Benthic cyanobacteria on coral reefs of Moorea Island (French Polynesia): diversity response to habitat quality. Hydrobiologia 843:61–78. https://doi.org/10.1007/s10750-019-04029-8
Acknowledgements
We would like to thank: Anne Hoggett and Lyle Vail at the Lizard Island Research Station, Paul Kench for help developing our coring technique and advice on coral reef taphonomy, Adrian Turner for assistance with microscopy and Howard Choat for ongoing support. Great thanks to Judy Sutherland and Wendy Nelson for advice on DNA extraction techniques for crustose coralline algae. Also thanks to Alessandro Pisaniello for practical guidance on DNA extraction.We acknowledge the use of New Zealand eScience Infrastructure (NeSI) high performance computing facilities.
Funding
Funding was provided by the University of Auckland School of Biological Sciences Performance Based Research Fund and University of Auckland Doctoral Scholarship.
Author information
Authors and Affiliations
Contributions
Both authors contributed to the study conception and design. GMN carried out the data collection and analysis and led the writing of the manuscript. KDC critically reviewed the versions of the manuscript. Both authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest in the writing of this study.
Ethics approval
Reef sampling was conducted under the Lizard Island Research Permit Granted by Great Barrier Reef Marine Park Authority (GBRMPA; Permit no. G14/36625.1). Fish were observed in their natural habitat and no fish were killed or injured during data collection for this study.
Additional information
Responsible Editor: W. Figueira.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: In this article Fig. 1 was wrongly appeared. It has been corrected.
Supplementary Information
Below is the link to the electronic supplementary material.
227_2023_4277_MOESM1_ESM.pdf
Supplementary file 1: Supplemental Fig. S1 Map of outer-shelf reef sites (Day and Hicks Reefs), adjacent to Lizard Island, Northern Great Barrier Reef, Australia, showing locations where the 120 bite cores from the six parrotfish species were extracted from the reef.
227_2023_4277_MOESM2_ESM.pdf
Supplementary file 2: Supplemental Fig. S2 Examples of in situ bite site photographs from Day and Hicks Reefs, showing bite marks (Photos KDC). Photographs were taken immediately after each bite was observed, prior to bite core extraction.
227_2023_4277_MOESM4_ESM.pdf
Supplementary file 4: Supplemental Fig. S5 Microscope photographs of Calothrix (Nostocales) from S. globiceps outer-shelf bite cores to highlight the presence of these cyanobacteria on S. globiceps cores, despite non-detection by 16S NGS molecular sequencing. Numbers represent S. globiceps outer-shelf core numbers.
227_2023_4277_MOESM5_ESM.pdf
Supplementary file 5: Supplemental Fig. S5 Microscope photographs of Calothrix (Nostocales) from S. globiceps outer-shelf bite cores to highlight the presence of these cyanobacteria on S. globiceps cores, despite non-detection by 16S NGS molecular sequencing. Numbers represent S. globiceps outer-shelf core numbers.
227_2023_4277_MOESM6_ESM.pdf
Supplementary file 6: Supplemental Fig. S6 Maximum-likelihood tree of bite core crustose coralline algae (CCA) ASVs from the 18S sequencing and reference sequences from SILVA 138. Phylogenetic tree was constructed with FASTTREE alignment and generated with iTOL. Clade support (bootstrap) values are shown with a black triangle. Coloured shaded circles show the presence of each ASV on the bite core sample for the six parrotfish species.
227_2023_4277_MOESM7_ESM.jpeg
Supplementary file 7: Supplemental Fig. S7 Ce. ocellatus feeding on a massive Porites at Mermaid Cove, Lizard Island, 2018 (pers obs. KDC).
227_2023_4277_MOESM8_ESM.docx
Supplementary file 8: Supplemental Table S1 Summary statistics from sequencing, including 3 letter code for the six parrotfish species.
227_2023_4277_MOESM9_ESM.pdf
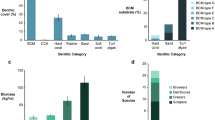
Supplementary file 9: Supplemental Table S2 Means and standard errors of percentage surface cover for all epilithic bite core biota as determined by CPCe on the outer-shelf.
227_2023_4277_MOESM10_ESM.docx
Supplementary file 10: Supplemental Table S3 Principal component analysis (PCA) on the bite core parameters Loadings over 0.3 and below -0.3 in BOLD. Percent of total variance for each principal component shown.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Nicholson, G.M., Clements, K.D. Fine-scale analysis of substrata grazed by parrotfishes (Labridae:Scarini) on the outer-shelf of the Great Barrier Reef, Australia. Mar Biol 170, 121 (2023). https://doi.org/10.1007/s00227-023-04277-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00227-023-04277-2