Abstract
Three strains of symbiotic bacteria were isolated from an entomopathogenic nematode Steinernema poinari retrieved from soil in eastern Poland. Using 16S rDNA, recA, gltX, gyrB, and dnaN gene sequences for phylogenetic analysis, these strains were shown to belong to the species Xenorhabdus bovienii. The nucleotide identity between the studied S. poinari microsymbionts and other X. bovienii strains calculated for 16S rDNA and concatenated sequences of four protein-coding genes was 98.7–100% and 97.9–99.5%, respectively. The phenotypic properties of the isolates also supported their close phylogenetic relationship with X. bovienii. All three tested X. bovienii strains of different Steinernema clade origin supported the recovery of infective juveniles and subsequent development of the nematode population. However, the colonization degree of new infective juvenile generations was significantly affected by the bacterial host donor/recipient. The colonization degree of infective juveniles reared on bacterial symbionts deriving from a non-cognate clade of nematodes was extremely low, but proved the possible host-switching between non-related Steinernema species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bacteria of the genus Xenorhabdus in symbiotic association with entomopathogenic nematodes (EPN) of the genus Steinernema Travassos (Nematoda: Steinermatidae) infect a wide range of soil-dwelling insects (Koppenhöfer and Gaugler 2009; Stock 2015). They have an unusual property of switching from a mutualistic to pathogenic lifestyle interacting with two different eukaryotic hosts and this triplicate (bacteria–nematode–insect) system can be easily established and investigated in the laboratory conditions. Bacteria of the genus Xenorhabdus are isolated from the intestinal lumen of their nematode host or infected insects as their natural habitats; they have never been found in free-living stage in the soil. Isolation and classification of EPN symbiotic bacteria were performed for the first time in 1964 and 26 species of the genus Xenorhabdus have been described to date (Table S1, supplementary material).
EPN and their microsymbionts have become a successful biological model facilitating investigation of prokaryote–eukaryote interactions (especially virulence mechanisms and a variety of bacterial natural products) as well as soil ecology (Campos-Herrera et al. 2012; Goodrich-Blair and Clarke 2007; Stock 2005). Additionally, both EPN and their bacterial symbionts play an important role in crop protection against insect pests, and identification of native nematode–bacteria associations is essential for successful control of pests in a particular area (Hiltpold 2015). Previously reported results indicated that most Xenorhabdus species are highly pathogenic to insects when directly injected without their nematode host (e.g. McMullan et al. 2017a; Sicard et al. 2004a, b; Sugar et al. 2012). This enhances the use of the bacteria (or only their genes) producing insecticidal toxins for biological control in agroforestry industries (Kumari et al. 2015; McMullen et al. 2017a; Zhang et al. 2012). Strains of some genus Xenorhabdus species can display a broad spectrum of genetic diversity. They differ in the virulence phenotype, which is determined by a wide variety of secreted bioactive components necessary for invasion, colonization, and use of the insect cadaver as a food source. Analyses of different Xenorhabdus strains increase the potential for discovery of novel natural products, including antibiotics, toxins, adhesins, hemolysins, proteases, and lipases (Hinchliffe et al. 2010; Murphin et al. 2015a). The increasing number of genus Xenorhabdus strains that are being sequenced is also a key factor in exploration thereof in medicine and pharmacy.
Although Xenorhabdus strains have the potential to become commercially important, the knowledge of the bacteria of the genus Xenorhabdus, especially their diversity within the species, is still insufficient. Many symbiotic bacteria of EPN have not been identified yet, which obscures comprehensive characterization of these organisms and limits their application.
The symbiotic Xenorhabdus–Steinernema association is specific, and each EPN species associates with only one Xenorhabdus species. However, most Xenorhabdus species display more flexibility in the nematode symbiont choice and may be associated with more than one Steinernema species (Stock 2015; Table S1, supplementary material). Previous studies have shown that X. bovienii bacteria can enter symbiosis with 14 EPN species, which makes such a broad host range a highly significant feature of this species. The EPN hosts of X. bovienii are distributed widely all over the world. They belong to two distinct clades of Steinernema distinguished by Nadler et al. (2006) on the basis of the sequences of nuclear and mitochondrial genes: affine–intermedium (clade I) and feltiae–kraussei (clade III). These nematodes constitute about half of the Steinernema species described so far.
Steinernema poinari was found for the first time in the Czech Republic, but it is quite abundant in Europe, also in Poland. Molecular and morphological characteristics have shown that this species is a member of the affine–intermedium group (Mráček et al. 2014). The genus Xenorhabdus bacteria described in this paper were isolated from three strains of S. poinari recovered in Poland. To reveal the taxonomic position of S. poinari microsymbionts, we analysed five loci 16S rDNA, recA, gltX, gyrB, and dnaN. The bacteria were also characterized phenotypically using biochemical and physiological tests. To date, there are no reports on S. poinari bacterial symbionts and this study is the first to provide information on their identification, phylogenetic relatedness, and phenotypic characteristics. We also performed experiments on the influence of different origin X. bovienii strains on EPN to uncover the possibility of symbiont exchange in this beneficial partnership.
Materials and methods
Bacterial isolation and phenotypic tests
The Xenorhabdus spp. strains used in this study were symbiotically associated with Polish isolates of S. poinari (Nematoda: Steinernematidae) (Table S2, supplementary material). All nematode strains were isolated from soil samples using the live trap method (Akhurst and Bedding 1975). Before isolation of the symbiotic bacteria, straight genetic lines of the nematodes were established [offspring of two infective juveniles (IJs)] and identified (microscopically and molecularly). The bacterial strains were isolated from the hemolymph of Galleria mellonella larvae (Lepidoptera: Pyralidae) as previously described (Kazimierczak et al. 2016). They were designated Xb041, Xb057, and Xb139 and are maintained in our laboratory collection. All biochemical and physiological tests were performed twice at 25 °C according to Kazimierczak et al. (2016).
Gene sequencing
Bacterial genomic DNA was extracted using a Genomic Mini AX Bacteria Spin Kit (A&A Biotechnology). 16S rDNA was amplified using primers 16SP1 and 16SP2 (Tailliez et al. 2006). The housekeeping genes coding the glutamyl-tRNA synthetase catalytic subunit (gltX), recombinase A (recA), DNA polymerase III beta chain (dnaN), and subunit B of DNA gyrase (gyrB) were amplified as described earlier (Kazimierczak et al. 2016). The PCR products were ligated into plasmids pJET1.2 (Thermo Fisher Scientific) according to the manufacturer’s instructions. The plasmids were transformed in E. coli XL1 Blue using a standard method. The inserts of positive clones were sequenced from both strands in Genomed (Poland).
Phylogenetic analysis
The 16S rRNA, gltX, recA, dnaN, and gyrB gene sequences of isolates Xb041, Xb057, and Xb139 obtained in this study were compared to GenBank nucleotide sequences using BLAST available on the NCBI website. Multiple sequence alignments were created using ClustalW at the default configuration. The evolutionary history of the studied Xenorhabdus strains was inferred using the neighbor-joining method in MEGA 6.06 (Tamura et al. 2004, 2013). The evolutionary distances were computed using the Tamura-Nei algorithm. All positions containing gaps and missing data were eliminated. To determine the statistical support for branches, bootstrapping with 1000 replicates of the data was conducted. There were 1334 positions in the final dataset for 16S rDNA, 382 for recA, 783 for dnaN, 1006 for gltX, and 811 for gyrB. The GeneBank accession numbers for the gene sequences specified in this study are as follows: MG995576-78 for 16S rDNA, MH001597-99 for recA, MH001600-02 for gyrB, MH001594-96 for dnaN, and MH001603-05 for gltX (Table S2, supplementary material).
Bacterial/EPN host interaction bioassays
We performed experiments on the influence of different origin symbiotic bacteria on (i) the recovery of IJs (termination of developmental diapause), (ii) nematode population development, and (iii) colonization degree of IJs (percent of IJs possessing a bacterial symbiont in their intestines). In these experiments, we used single bacterial strains symbiotically associated with S. poinari (strain Xb057), S. affine (strain Xb-aff), S. intermedium (strain Xb-int) (the same nematode clade), S. feltiae (strain Xb-fel), and S. silvaticum (strain Xb-Z1Z) (another Steinernema clade symbiotically associated with X. bovienii). The nematodes were reared on Petri dishes with modified Wouts agar (1981) [1% (w/v) full-fat soy flour, 1% (w/v) lyophilized yeast, 1% (w/v) dried egg yolk, 2% (w/v) agar, 0.5% (w/v) NaCl, 1000 ml deionized water]. After 48 h of bacterial growth at 25 °C, the plates were inoculated with ~ 100 of surface-sterilized IJs [three 15-min washes in a 0.4% (w/v) Hyamine® solution, two 5-min washes in sterile deionized water]. The plates with the nematodes were placed on modified White traps (Stock and Goodrich-Blair 2012) at 17.5 °C and were regularly checked. After 2–3 weeks, new generations of nematodes were harvested from White traps and stored in tap water at 12 °C. The colonization degree of IJs was determined for groups of 20 IJs and insects separately, dividing the number of dead insects by the number of infected insects (dead and live) as previously described (Kazimierczak et al. 2018). For each combination of the bacteria and nematode, the colonization degree was determined for five groups of IJs and insects. All experiments were repeated twice with the use of nematodes and G. mellonella larvae originating from different batches.
Statistical analysis
Since there was no effect of the experiment replication on the nematode colonization degree, the data from the two replications of the experiment were combined for the analysis. Before analysis, the data were normalized by arcsin square root transformation. The normality of data distribution was determined with the Shapiro–Wilk test and homogeneity of variance was assessed with Levene’s test. Analysis of variance and means separation with Tukey HSD test were used for comparisons. Back-transformed means ± SE are presented. All analyses were conducted using SPSS Statistics 24 software (IBM).
Results and discussion
Phylogeny of the S. poinari microsymbionts based on 16S rDNA
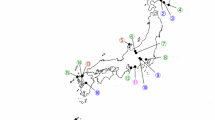
The use of the 16S rRNA gene has proved useful for classification, identification, and characterization of entomopathogenic nematode bacteria. Nevertheless, many studies have shown such confounding factors in 16S rRNA phylogeny as evidence of lateral gene transfer (LTG) and a low level of variation between 16S rDNA sequences, including Xenorhabdus spp. (Lee and Stock 2010a; Tailliez et al. 2006). It is generally accepted that 16S rRNA gene sequence similarity of two strains < 98.7 indicates that they belong to different species, but there is no defined threshold of 16S rDNA similarity above which it is possible to identify bacteria at a species level (Stackebrandt and Ebers 2006; Yarza et al. 2014). The sequences of 16S rDNA obtained for Xb041, Xb057, and Xb139 isolates shared 99.8–100% sequence similarity with each other (maximum 3 bp differences), which placed all the isolates in the same species. The analysed sequences were most similar to the 16S rDNA sequences reported for X. bovienii displaying 98.7–100% identity with them (max. 16 nucleotide substitutions). The phylogenetic analysis of the 16S rDNA sequences divided the X. bovienii strains and our isolates into two groups. All of the Polish S. poinari isolates were placed in a large cluster together with X. bovienii strains being symbiotic partners of nematodes from both groups feltiae–kraussei and affine–intermedium (Fig. 1).
Neighbor-joining tree showing the phylogenetic relationships of the S. poinari microsymbionts studied (bolded) with X. bovienii strains based on 16S rRNA gene sequences. Bootstrap values based on 1000 replicates > 70% are indicated at the branching points. The scale bar presents the number of nucleotide substitutions per site. The sequences of Photorhabdus luminescens subsp. laumondii and Photorhabdus asymbiotica subsp. asymbiotica were used as an outgroup
Phylogeny of the X. bovienii isolates based on the combination of recA, gltX, gyrB, and dnaN gene fragments
Taking into account the conservative nature of 16S rDNA sequences between X. bovienii strains, additional phylogenetic information was derived from the analysis of four housekeeping gene sequences. This multi-gene approach increases the discriminatory power of phylogenetic analysis and prevents misclassification of new isolates. In particular, comparative sequence analysis of recA, gltX, gyrB, and dnaN has been widely used for the diagnostics of entomopathogenic bacteria species (e.g. Dreyer et al. 2017; Ferreira et al. 2013; Tailliez et al. 2012). On the basis of grouping obtained with recA, gyrB, dnaN, and gltX gene sequences, Tailliez et al. (2010) have demonstrated that all X. bovienii strains constitute a separate clade within the genus Xenorhabdus. This broad phylogenetic study has provided evidence that the genus Xenorhabdus strains that shared less than 97% nucleotide identity of the concatenated sequences of the recA, gyrB, dnaN and gltX do not belong to the same species. In our analysis, concatenated 2990-bp-long sequences of these genes for the Polish isolates of S. poinari shared 99.2–99.8% similarity with each other and 97.9–99.5 with those reported for X. bovienii. In the phylogram constructed on the basis of the concatenated sequences of recA, gltX, gyrB, and dnaN genes (Fig. 2), all the Polish S. poinari isolates formed a homogeneous and highly supported cluster with other X. bovienii strains. This confirms their identification as X. bovienii.
Neighbor-joining tree showing the phylogenetic relationships of S. poinari microsymbionts studied (bolded) with X. bovienii strains based on concatenated recA, gyrB, dnaN, and gltX gene sequences. Bootstrap values based on 1000 replicates > 70% are indicated at the branching points. The scale bar presents the number of nucleotide substitutions per site. The sequence of Photorhabdus asymbiotica subsp. asymbiotica was used as an outgroup
It is known that the sequences of recA, gltX, gyrB, and dnaN genes within the genus Xenorhabdus are highly conserved and display similarity > 80% (Tailliez et al. 2010). The weak phylogenetic signal of each individual housekeeping gene usually resulted in low bootstrap values and a high incidence of unresolved branches in the evolutionary analysis of Xenorhabdus (Lee and Stock 2010a). To assess the amount of phylogenetic information associated with each protein-coding gene analysed, we determined the number of variable and parsimony-informative sites in single-gene alignments. Gene gltX had the highest number of parsimony-informative positions among the analysed X. bovienii strains − 44 (4.4%), and the lowest number was recorded for recA − 7 positions (1.8%) and gyrB − 13 (1.6%). The studied Polish S. poinari microsymbionts showed a high level of gyrB, dnaA, and recA gene conservation—maximum double and single synonymous substitution was detected for gyrB and dnaN, respectively, whereas the recA gene sequences were identical. In contrast, the gltX gene in the studied isolates was more divergent—strain Xb041 differed from Xb057 and Xb139 by 17 and 14 positions, respectively. Interestingly, Xb041 displayed 100% similarity in the gltX gene sequence to X. bovienii GE02, a bacterial symbiont of S. tbilisiensis. This resulted in grouping together of the Xb041 isolate and the X. bovienii GE02 strain on the gltX tree in a distant position from the other bacteria included in the analysis (Fig. S1). This relationship was not revealed by the phylogenetic analysis of the 16S rDNA, recA, and dnaN; according to the sequences of these loci, our isolates form a homogenous group with the X. bovienii strains (Fig. 1, Fig. S3, Fig. S4). In turn, the gyrB gene of the studied isolates displayed relatively high similarity to the X. bovienii GE02 gyrB gene (1–3 substitutions), resulting in their placement in the same clade on the phylogram (Fig. S2). The discrepancies in individual gene trees may be a result of LTG or different evolutionary pressures on the gene studied. Despite the high resistance of housekeeping genes to LTG, the occurrence of recombination for gltX, gyrB, and serC within the Xenorhabdus clade is known (Sergeant et al. 2006; Tailliez et al. 2010). It supports the assumption that the ambiguous phylogenetic relationships among the studied strains could be caused by lateral gene transfer.
Phenotypic characterization of bacterial isolates
All the tested strains of the genus Xenorhabdus were Gram-negative, catalase-, urease- and cytochrome oxidase-negative rods. The bacterial strains used in our study absorbed bromothymol blue from NBTA and neutral red from MacConkey agar plates. The Xenorhabdus colonies growing on the LBA plates were yellow and the size of bacterial cells was variable (2.4–8.3 µm). The maximum growth temperature on the LBA plates was in the range of 32–33 °C. The tested bacterial strains were motile, ampicillin resistant, and able to β-haemolyse on sheep blood agar and hydrolyse casein and gelatin. The Xenorhabdus isolates associated with S. poinari were arginine dihydrolase, DNase, and phospholipase positive. All the strains metabolized N-acetyl-d-glucosamine, α-d-glucose, d-mannose, d-trehalose, d-gluconic acid, inosine, uridine, glucose-1-phosphate, and glucose-6-phosphate in aerobic conditions. Tween®80, methyl pyruvate, p-hydroxy-phenylacetic acid, d,l-lactic acid, l-alanine, l-alanyl-glycine, l-asparagine, l-histidine, glycerol and d,l-α-glycerol phosphate were utilized weakly. The bacterial strains used in this study fermented d-glucose, d-mannose, and N-acetylglucosamine. d-Trehalose, potassium gluconate, and potassium-5-ketogluconate were fermented weakly. The results of the other oxidation and fermentation tests were strain dependent (compare Table 1). All the tested strains of the genus Xenorhabdus exhibited phenotypic characteristics typical of X. bovienii bacteria (Boemare and Akhurst 1988; Tailliez et al. 2006), which confirms their molecular identification.
Influence of different X. bovienii strains on the entomopathogenic nematodes
All the tested strains of X. bovienii supported the recovery of IJs and subsequent development of the nematode population. However, the colonization degree of the new IJ generations was significantly affected by the bacterial host donor/recipient (F4,45≥617.76, P < 0.001, Fig. 3). There was no significant effect of the X. bovienii strains on the colonization degree of harvested IJs when the bacterial donor and recipient were derived from the same clade of nematodes. In these cases, the colonization degree of IJs was ~ 80% for the affine–intermedium clade and > 95% for the feltiae–kraussei clade. When the nematodes were reared on bacteria from another clade of Steinernema, the mean colonization degree of harvested IJs was low (0.5/2.7%).
Effect of native- and non-native bacteria on the mean (± SE) colonization degree of Steinernema spp. IJs. Columns with the same letter are not significantly different within the bacterial host donor (P < 0.05, Tukey HSD test). S. aff—S. affine, S. fel—S. feltiae, S. int—S. intermedium, S. poi—S. poinari, S. sil—S. silvaticum
Phylogenetic data demonstrate that strains of X. bovienii are often grouped together on phylogenetic trees regardless of the phylogenetic origin of their nematode host. For example, the X. bovienii Si and SS-2004 strains are derived from phylogenetically distant hosts (S. intermedium from clade I and S. jollieti from clade III, respectively), but they occupy the same place in the phylogenetic trees. A similar trend has been noted in the tested Xb041, Xb057, and Xb139 microsymbionts of S. poinari from clade I—they are phylogenetically very close to the strains of X. bovienii isolated from Steinernema spp. belonging to clade III (Figs. 1, 2). These results suggest a low level of specificity in the symbiotic interaction of X. bovienii and their Steinernema partners and strengthen the hypothesis of strong host-switching shaping these relationships. This is in agreement with the previous observation of host switches in the case of X. bovienii, a symbiont of S. affine and S. intermedium (Lee and Stock 2010b). The ability to switch between nematodes from distantly related clades has also been reported for X. khoisanae or X. nematophila bacteria (Dreyer et al. 2017; Stock 2015). However, a cophylogenetic study of 30 Steinernema/Xenorhabdus pairs revealed 17 apparent mismatches between two trees, indicating host-switching as well as 12 cospeciation events. This suggests that a high level of specificity between the nematode host and the bacterial partner can occur for some host–symbiont phylogenetic relationships (Lee and Stock 2010b). Similarly, another cophylogenetic analysis has indicated specificity and coevolution in symbiotic partner pairs as evidenced by remarkable congruence between the phylogenies of nematodes from the bicornutum group of Steinernema and their symbiotic bacteria (Bhat et al. 2018) as well as between S. feltiae and S. puntauvense/X. bovienii associations (Murfin et al. 2015b). A high level of specificity with limited horizontal transfers between these symbiotic partners was also suggested in a study conducted by Emelianoff et al. (2008), which showed limited transfers of X. bovienii bacteria between S. feltiae and S. affine living in the same area.
The results of the switches of Xenorhabdus between different Steinernema species in laboratory experiments also vary depending on the pairs considered. However, the experimental tests revealed that closely related bacterial strains provide nematodes with greater virulence than more phylogenetically divergent ones. The reproductive fitness of both symbionts decreased as the phylogenetic distance between native and non-native nematode partner increased (McMullen et al. 2017b; Murfin et al. 2015b).
Symbiosis of numerous nematode species with X. bovienii can reduce interspecific competition between Steinernema species, increasing the size of potential food resources. It can also explain the worldwide occurrence and frequent coexistence of affine–intermedium and feltiae–kraussei Steinernema clades (Půža and Mráček 2010) and probably frequent host-switching events (Murfin et al. 2015b). However, the results of our study indicate that when the donors and recipients of X. bovienii represent a different nematode clade, the colonization degree of newly formed IJs is very low. This proves that host-switching events are rare, but possible. Previously published studies (Grewal et al 1997; Sicard et al. 2004a, b) have shown that secondary infection of insects by nematodes from another Steinernema clade leads to termination of nematode diapause, their population development, and formation of new IJs. These new IJs, as we have shown, have much lower competitive abilities because most of them are axenic. Axenic IJs are able to infect a new host, but without symbiotic bacteria they are not able to reproduce and fully utilize food resources. Host co-infection or secondary infection by nematodes from different Steinernema clades symbiotically associated with X. bovienii may, therefore, lead to reduction of their competitive abilities. The mechanisms of the coexistence of competitor populations remain unknown and will be investigated.
Conclusions
We found that entomopathogenic nematodes S. poinari are symbiotically associated with X. bovienii. This result increases the known range of X. bovienii hosts and confirms that nematodes from the affine–intermedium and feltiae–kraussei clades of Steinernema enter symbiosis with X. bovienii exclusively. All three strains of X. bovienii used in our experiments supported the recovery of IJs and development of the nematode population. The colonization degree of IJs reared on bacterial symbionts deriving from a non-cognate clade of nematodes was extremely low, but proved the possible host-switching of X. bovienii between non-related Steinernema species.
Abbreviations
- IJ (s):
-
Infective juvenile (s)
- EPN:
-
Entomopathogenic nematode (s)
- BLAST:
-
Basic Local Alignment Search Tool
- NCBI:
-
National Center for Biotechnology Information
- w/v:
-
Weight per volume
- NBTA:
-
Nutrient bromothymol blue agar
- LBA:
-
Luria–Bertani agar
- SE:
-
Standard error
- LTG:
-
Lateral gene transfer
References
Akhurst RJ (1980) Morphological and functional dimorphism in Xenorhabdus spp., bacteria symbiotically associated with the insect pathogenic nematodes Neoaplectana and Heterorhabditis. Microbiol 121(2):303–309. https://doi.org/10.1099/00221287-121-2-303
Akhurst RJ (1983) Taxonomic study of Xenorhabdus, a genus of bacteria symbiotically associated with insect pathogenic nematodes. Int J Syst Bacteriol 33(1):38–45. https://doi.org/10.1099/00207713-33-1-38
Akhurst RJ, Boemare NE (1988) A numerical taxonomic study of the genus Xenorhabdus (Enterobacteriaceae) and proposed elevation of the subspecies of X. nematophilus to species. J Gen Microbiol 134(7):1835–1845. https://doi.org/10.1099/00221287-134-7-1835
Bedding RA, Akhurst RJ (1975) A simple technique for the detection of insect parasitic rhabditid nematodes in soil. Nematologica 21(1):10–110. https://doi.org/10.1163/187529275X00419
Bhat AH, Chaubey AK, Puza V (2018) The first report of Xenorhabdus indica from Steinernema pakistanense. Co-phylogenetic study suggests co-speciation between X. indica and its steinernematid nematodes. J Helminthol. https://doi.org/10.1017/S0022149X17001171
Boemare NE, Akhurst RJ (1988) Biochemical and physiological characterization of colony form variants in Xenorhabdus spp. (Enterobacteriaceae). J Gen Microbiol 134:751–761. https://doi.org/10.1099/00221287-134-3-751
Campos-Herrera R, Barbercheck M, Hoy CW, Stock SP (2012) Entomopathogenic nematodes as a model system for advancing the frontiers of ecology. J Nematol 44(2):162–176
Dreyer J, Malan AP, Dicks LMT (2017) Three novel Xenorhabdus-Steinernema associations and evidence of strains of X. khoisanae switching between different clades. Curr Microbiol 74(8):938–942. https://doi.org/10.1007/s00284-017-1266-2
Emelianoff V, Le Brun N, Pages S, Stock SP, Tailliez P, Moulia C, Sicard M (2008) Isolation and identification of entomopathogenic nematodes and their symbiotic bacteria from Herault and Gard (Southern France). J Invertebr Pathol 98(2):211–217. https://doi.org/10.1016/j.jip.2008.01.006
Ferreira T, van Reenen CA, Endo A, Sproer C, Malan AP, Dicks LMT (2013) Description of Xenorhabdus khoisanae sp. nov., the symbiont of the entomopathogenic nematode Steinernema khoisanae. Int J Syst Evol Micr 63(9):3220–3224. https://doi.org/10.1099/ijs.0.049049-0
Goodrich-Blair H, Clarke DJ (2007) Mutualism and pathogenesis in Xenorhabdus and Photorhabdus. Two roads to the same destination. Mol Microbiol 64(2):260–268. https://doi.org/10.1111/j.1365-2958.2007.05671.x
Grewal PS, Lewis EE, Gaugler R (1997) Response of infective stage parasites (Nematoda: Steinernematidae) to volatile cues from infected hosts. J Chem Ecol 23(2):503–515
Hiltpold I (2015) Prospects in the application technology and formulation of entomopathogenic nematodes for biological control of insect pests. In: Campos-Herrera R (ed) Nematode pathogenesis of insects and other pests: ecology and applied technologies for sustainable plant and crop protection. Springer, Neuchâtel, pp 187–206. https://doi.org/10.1007/978-3-319-18266-7_1
Hinchliffe SJ, Hares M, Dowling A, ffrench-Constant R (2010) Insecticidal toxins from the Photorhabdus and Xenorhabdus bacteria. Open Toxinol J 3:83–100. https://doi.org/10.2174/1875414701003010101
Kämpfer P, Tobias NJ, Ke LP, Bode HB, Glaeser SP (2017) Xenorhabdus thuongxuanensis sp. nov. and Xenorhabdus eapokensis sp. nov., isolated from Steinernema species. Int J Syst Evol Microbiol 67(5):1107–1114. https://doi.org/10.1099/ijsem.0.001770
Kazimierczak W, Sajnaga E, Skowronek M, Kreft AM, Skrzypek HW, Wiater A (2016) Molecular and phenotypic characterization of Xenorhabdus bovienii symbiotically associated with Steinernema silvaticum. Arch Microbiol 198(10):995–1003. https://doi.org/10.1007/s00203-016-1261-1
Kazimierczak W, Lis M, Skrzypek T, Kreft A (2018) Comparison of the methods applicable for the pathogenicity assessment of entomopathogenic nematodes. Biocontrol 63(2):289–298. https://doi.org/10.1007/s10526-017-9856-2
Koppenhöfer HS (2007) Bacterial symbionts of Steinernema and Heterorhabditis. In: Nguyen KB, Hunt DJ (eds) Entomopathogenic nematodes: systematics, phylogeny and bacterial symbionts, Nemathology Monographs and Perspectives, 5. Brill Academic Publishers, Boston, pp 735–808
Koppenhöfer H, Gaugler R (2009) Entomopathogenic nematode and bacteria mutualism. In: White J, Torres M (eds) Defensive mutualism in microbial symbiosis. CRC Press, Boca Raton, pp 99–116
Kumari P, Mahapatro GK, Banerjee N, Sarin NB (2015) Ectopic expression of GroEL from Xenorhabdus nematophila in tomato enhances resistance against Helicoverpa armigera and salt and thermal stress. Transgenic Res 24(5):859–873. https://doi.org/10.1007/s11248-015-9881-9
Kuwata R, Qiu L-H, Wang W, Harada Y, Yoshida M, Kondo E, Yoshiga T (2013) Xenorhabdus ishibashii sp. nov., isolated from the entomopathogenic nematode Steinernema aciari. Int J Syst Evol Microbiol 63(5):1690–1695. https://doi.org/10.1099/ijs.0.041145-0
Lee M-M, Stock SP (2010a) A multigene approach for assessing evolutionary relationships of Xenorhabdus spp. (gamma-Proteobacteria), the bacterial symbionts of entomopathogenic Steinernema nematodes. J Invertebr Pathol 104(2):67–74. https://doi.org/10.1016/j.jip.2010.01.005
Lee M-M, Stock SP (2010b) A multilocus approach to assessing co-evolutionary relationships between Steinernema spp. (Nematoda. Steinernematidae) and their bacterial symbionts Xenorhabdus spp. (γ-Proteobacteria: Enterobacteriaceae) Syst Parasitol 77(1):1–12. https://doi.org/10.1007/s11230-010-9256-9
Lengyel K, Lang E, Fodor A, Szallas E, Schumann P, Stackebrandt E (2005) Description of four novel species of Xenorhabdus, family Enterobacteriaceae: Xenorhabdus budapestensis sp. nov., Xenorhabdus ehlersii sp. nov., Xenorhabdus innexi sp. nov., and Xenorhabdus szentirmaii sp. nov. Syst Appl Microbiol 28(2):115–122. https://doi.org/10.1016/j.syapm.2004.10.004
McMullen JG, McQuade R, Ogier J-C, Pages S, Gaudriault S, Stock SP (2017a) Variable virulence phenotype of Xenorhabdus bovienii (gamma-Proteobacteria. Enterobacteriaceae) in the absence of their vector hosts. Microbiology 163(4):510–522. https://doi.org/10.1099/mic.0.000449
McMullen JG, Peterson BF, Forst S, Goodrich-Blair H, Stock SP (2017b) Fitness costs of symbiont switching using entomopathogenic nematodes as a model. BMC Evol Biol 17(1):100. https://doi.org/10.1186/s12862-017-0939-6
Mráček Z, Nermut J (2014) Steinernema poinari sp. n. (Nematoda: Steinernematidae) a new entomopathogenic nematode from the Czech Republic. Zootaxa 3760:336–350. https://doi.org/10.11646/zootaxa.3760.3.2
Murfin KE, Whooley AC, Klassen JL, Goodrich-Blair H (2015a) Comparison of Xenorhabdus bovienii bacterial strain genomes reveals diversity in symbiotic functions. BMC Genom 16:889. https://doi.org/10.1186/s12864-015-2000-8
Murfin KE, Lee M-M, Klassen JL, McDonald BR, Larget B, Forst S, Stock SP, Currie CR, Goodrich-Blair H (2015b) Xenorhabdus bovienii strain diversity impacts coevolution and symbiotic maintenance with Steinernema spp. nematode hosts. mBio 6(3):e00076. https://doi.org/10.1128/mBio.00076-15
Nadler SA, Bolotin E, Stock SP (2006) Phylogenetic relationships of Steinernema Travassos, 1927 (Nematoda. Cephalobina: Steinernematidae) based on nuclear, mitochondrial and morphological data. Syst Parasitol 63(3):161–181. https://doi.org/10.1007/s11230-005-9009-3
Nishimura Y, Hagiwara A, Suzuki T, Yamanaka S (1994) Xenorhabdus japonicus sp. nov. associated with the nematode Steinernema kushidai. World J Microb Biot 10(2):207–210. https://doi.org/10.1007/BF00360889
Poinar GO, Thomas GM (1965) A new bacterium, Achromobacter nemathopilus sp. nov. (Achromobacteriaceae: Eubacteriales), associated with a nematode. Int B Bact Nomencl T 15:249–252
Půža V, Mráček Z (2010) Mechanism of coexistence of two sympatric entomopathogenic nematodes, Steinernema affine and S. kraussei (Nematoda: Steinernematidae) in a central European oak woodland soil. Appl Soil Ecol 45:65–70. https://doi.org/10.1016/j.apsoil.2010.02.002
Sergeant M, Baxter L, Jarrett P, Shaw E, Ousley M, Winstanley C, Morgan JAW (2006) Identification, typing, and insecticidal activity of Xenorhabdus isolates from entomopathogenic nematodes in United Kingdom soil and characterization of the xpt toxin loci. Appl Environ Microb 72(9):5895–5907. https://doi.org/10.1128/AEM.00217-06
Sicard M, Brugirard-Ricaud K, Pages S, Lanois A, Boemare NE, Brehelin M, Givaudan A (2004a) Stages of infection during the tripartite interaction between Xenorhabdus nematophila, its nematode vector, and insect hosts. App Environ Microb 70(11):6473–6480. https://doi.org/10.1128/AEM.70.11.6473-6480.2004
Sicard M, Ferdy JB, Pages S, Le Brun N, Godelle B, Boemare N, Moulia C (2004b) When mutualists are pathogens: an experimental study of the symbioses between Steinernema (entomopathogenic nematodes) and Xenorhabdus (bacteria). J Evol Biol 17(5):985–993. https://doi.org/10.1111/j.1420-9101.2004.00748.x
Somvanshi VS, Lang E, Ganguly S, Swiderski J, Saxena AK, Stackebrandt E (2006) A novel species of Xenorhabdus, family Enterobacteriaceae: Xenorhabdus indica sp. nov., symbiotically associated with entomopathogenic nematode Steinernema thermophilum Ganguly and Singh, 2000. Syst Appl Microbiol 29(7):519–525. https://doi.org/10.1016/j.syapm.2006.01.004
Stackebrandt E, Ebers J (2006) Taxonomic parameters revised: tarnished gold standards. Microbiol Today 33(4):152–155
Stock SP (2005) Insect-parasitic nematodes. From lab curiosities to model organisms. J Invertebr Pathol 89(1):57–66. https://doi.org/10.1016/j.jip.2005.02.011
Stock SP (2015) Diversity, biology and evolutionary relationships. In: Campos-Herrera R (ed) Nematode pathogenesis of insects and other pests: ecology and applied technologies for sustainable plant and crop protection. Springer, Neuchâtel, pp 3–27. https://doi.org/10.1007/978-3-319-18266-7_1
Stock SP, Goodrich-Blair H (2012) Nematode parasites, pathogens and associates of insects and invertebrates of economic importance. In: Lacey LA (ed) Manual of techniques in invertebrate pathology. Elsevier, New York, pp 373–426. https://doi.org/10.1016/B978-0-12-386899-2.00012-9
Sugar DR, Murfin KE, Chaston JM, Andersen AW, Richards GR, deLeon L, Baum JA, Clinton WP, Forst S, Goldman BS, Krasomil-Osterfeld KC, Slater S, Stock SP, Goodrich-Blair H (2012) Phenotypic variation and host interactions of Xenorhabdus bovienii SS-2004, the entomopathogenic symbiont of Steinernema jollieti nematodes. Environ Microbiol 14(4):924–939. https://doi.org/10.1111/j.1462-2920.2011.02663.x
Tailliez P, Pages S, Ginibre N, Boemare N (2006) New insight into diversity in the genus Xenorhabdus, including the description of ten novel species. Int J Syst Evol Micr 56(12):2805–2818. https://doi.org/10.1099/ijs.0.64287-0
Tailliez P, Laroui C, Ginibre N, Paule A, Pages S. Boemare N (2010) Phylogeny of Photorhabdus and Xenorhabdus based on universally conserved protein-coding sequences and implications for the taxonomy of these two genera. Proposal of new taxa. X. vietnamensis sp. nov. P. luminescens subsp. caribbeanensis subsp. nov., P. luminescens subsp. hainanensis subsp. nov., P. temperata subsp. khanii subsp. nov., P. temperata subsp. tasmaniensis subsp. nov., and the reclassification of P. luminescens subsp. thracensis as P. temperata subsp. thracensis comb. nov. Int J Syst Evol Micr 60(8):1921–1937. https://doi.org/10.1099/ijs.0.014308-0
Tailliez P, Pages S, Edgington S, Tymo LM, Buddie AG (2012) Description of Xenorhabdus magdalenensis sp. nov., the symbiotic bacterium associated with Steinernema australe. Int J Syst Evol Micr 62(8):1761–1765. https://doi.org/10.1099/ijs.0.034322-0
Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbour-joining method. P Natl Acad Sci USA 101(30):11030–11035. https://doi.org/10.1073/pnas.0404206101
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Wouts WM (1981) Mass production of the entomogenous nematode Heterorhabditis heliothidis (Nematoda: Heterorhabditidae) on artificial media. J Nematol 13(4):467–469
Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, Schleifer K-H, Whitman WB, Euzéb J, Amann R, Rosselló-Móra R (2014) Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol 12(9):635–645. https://doi.org/10.1038/nrmicro3330
Zhang H, Mao J, Liu F, Zeng F (2012) Expression of a nematode symbiotic bacterium-derived protease inhibitor protein in tobacco enhanced tolerance against Myzus persicae.. Plant Cell Rep 31(11):1981–1989. https://doi.org/10.1007/s00299-012-1310-4
Acknowledgements
This work was supported by the Polish Ministry of Science and Higher Education grant for statutory activity. The authors gratefully acknowledge the use of the services and facilities of the Centre for Interdisciplinary Research of the John Paul II Catholic University of Lublin, Lublin, Poland, co-funded by the European Union from the European Regional Development Fund under the Operational Program Development of Eastern Poland 2007–2013 (POPW.01.03.00-06-003/09-00).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Erko Stackebrandt.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Sajnaga, E., Kazimierczak, W., Skowronek, M. et al. Steinernema poinari (Nematoda: Steinernematidae): a new symbiotic host of entomopathogenic bacteria Xenorhabdus bovienii. Arch Microbiol 200, 1307–1316 (2018). https://doi.org/10.1007/s00203-018-1544-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-018-1544-9