Abstract
Key message
The Mendelian gene conferring resistance to Soybean mosaic virus Strain SC20 in soybean was fine-mapped onto a 79-kb segment on Chr.13 where two closely linked candidate genes were identified and qRT-PCR verified.
Abstract
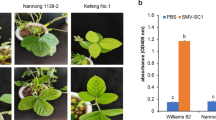
Soybean mosaic virus (SMV) threatens the world soybean production, particularly in China. A country-wide SMV strain system composed of 22 strains was established in China, among which SC20 is a dominant strain in five provinces in Southern China. Resistance to SC20 was evaluated in parents, F1, F2 and the F2:7 RIL (recombinant inbred line) population derived from a cross between Qihuang-1 (resistant) and NN1138-2 (susceptible). The segregation ratio of resistant to susceptible in the populations suggested a single dominant gene involved in the resistance to SC20 in Qihuang-1. A “partial genome mapping strategy” was used to map the resistance gene on Chromosome 13. Linkage analysis between 178 RILs and genetic markers showed that the SC20-resistance gene located at 3.9 and 3.8 cM to the flanking markers BARCSOYSSR_13_1099 and BARCSOYSSR_13_1185 on Chromosome 13. Subsequently, a residual heterozygote segregating population with 346 individuals was developed by selfing four plants heterozygous at markers adjacent to the tentative SC20-resistance gene; then, the candidate region was delimited to a genomic interval of approximately 79 kb flanked by the new markers gm-ssr_13-14 and gm-indel_13-3. Among the seven annotated candidate genes in this region, two genes, Glyma.13G194700 and Glyma.13G195100, encoding Toll Interleukin Receptor–nucleotide-binding–leucine-rich repeat resistance proteins were identified as candidate resistance genes by quantitative real-time polymerase chain reaction and sequence analysis. The two closely linked genes work together to cause the phenotypic segregation as a single Mendelian gene. These results will facilitate marker-assisted selection, gene cloning and breeding for the resistance to SC20.







Similar content being viewed by others
References
Arif M, Hassan S (2002) Evaluation of resistance in soybean germplasm to Soybean mosaic potyvirus under field conditions. J Biol Sci 2:601–604
Ashfield T, Danzer J, Held D, Clayton K, Keim P, Maroof MS, Webb D, Innes R (1998) Rpg1, a soybean gene effective against races of bacterial blight, maps to a cluster of previously identified disease resistance genes. Theor Appl Genet 96:1013–1021
Ashfield T, Ong LE, Nobuta K, Schneider CM, Innes RW (2004) Convergent evolution of disease resistance gene specificity in two flowering plant families. Plant Cell 16:309–318
Bai L, Li HC, Ma Y, Wang DG, Liu N, Zhi HJ (2009) Inheritance and gene mapping of resistance to soybean mosaic virus strain SC-11 in soybean. Soybean Sci 28:1–6
Bryan GT, Wu KS, Farrall L, Jia Y, Hershey HP, McAdams SA, Faulk KN, Donaldson GK, Tarchini R, Valent B (2000) A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell 12:2033–2046
Chen ML, Luo J, Shao GN, Wei XJ, Tang SQ, Sheng ZH, Song J, Hu PS (2012) Fine mapping of a major QTL for flag leaf width in rice, qFLW4, which might be caused by alternative splicing ofNAL1. Plant Cell Rep 31:863–872
Cheng HJ, Guo SG, Yu ZL (1982) Major diseases and pests of soybeans in China proceedings of the first China/USA soybean symposium and working group meeting, pp 74–80, 26–30 July 1982
Diers B, Mansur L, Imsande J, Shoemaker R (1992) Mapping Phytophthora resistance loci in soybean with restriction fragment length polymorphism markers. Crop Sci 32:377–383
Dodds PN, Lawrence GJ, Ellis JG (2001) Six amino acid changes confined to the leucine-rich repeat β-strand/β-turn motif determine the difference between the P and P2 rust resistance specificities in flax. Plant Cell 13:163–178
Doyle JJ (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Dutta S, Kumawat G, Singh BP, Gupta DK, Singh S, Dogra V, Gaikwad K, Sharma TR, Raje RS, Bandhopadhya TK (2011) Development of genic-SSR markers by deep transcriptome sequencing in pigeonpea [Cajanus cajan (L.) Mills paugh]. BMC Plant Biol 11:1–13
Fu SX, Zhan Y, Zhi HJ, Gai JY, Yu DY (2006) Mapping of SMV resistance gene Rsc7 by SSR markers in soybean. Genetica 128:63–69
Gore MA, Hayes AJ, Jeong SC, Yue YG, Buss GR, Saghai Maroof MA (2002) Mapping tightly linked genes controlling potyvirus infection at the Rsv1 and Rpv1 region in soybean. Genome 45:592–599
Guo D, Zhi H, Wang Y, Gai J, Zhou X, Yang C (2005) Identification and distribution of strains of Soybean mosaic virus in middle and northern of Huang Huai Region of China. Soybean Sci 27:64–68
Guo DQ, Wang YW, Zhi HJ, Gai JY, Li HC, Li K (2007) Inheritance and gene mapping of resistance to SMV strain group SC-13 in soybean. Soybean Sci 26:21–24
Hayes A, Jeong S, Gore M, Yu Y, Buss G, Tolin S, Maroof MS (2004) Recombination within a nucleotide-binding-site/leucine-rich-repeat gene cluster produces new variants conditioning resistance to soybean mosaic virus in soybeans. Genetics 166:493–503
Hirata K, Masuda R, Tsubokura Y, Yasui T, Yamada T, Takahashi K, Nagaya T, Sayama T, Ishimoto M, Hajika M (2014) Identification of quantitative trait loci associated with boiled seed hardness in soybean. Breed Sci 64:362–370
Jeong S, Maroof S (2004) Detection and genotyping of SNPs tightly linked to two disease resistance loci, Rsv1 and Rsv3, of soybean. Plant Breeding 123:305–310
Jeong SC, Kristipati S, Hayes AJ, Maughan PJ, Noffsinger SL, Gunduz I, Bussa GR, Saghai Maroof MA (2002) Genetic and sequence analysis of markers tightly linked to the Soybean mosaic virus resistance gene, Rsv3. Crop Sci 42:265–270
Jiang S, Zhang X, Zhang F, Jiang S, Zhang X, Zhang F, Xu Z, Chen W, Li Y (2012) Identification and fine mapping of qCTH4, a quantitative trait loci controlling the chlorophyll content from tillering to heading in rice (Oryza sativa L.). J Hered 103:720–726
Kosambi DD (1943) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Li HC, Zhi HJ, Gai JY, Guo DQ, Wang YW, Li K, Bai L, Yang H (2006) Inheritance and gene mapping of resistance to soybean mosaic virus strain SC14 in soybean. J Integr Plant Biol 48:1466–1472
Li K, Ren R, Karthikeyan A, Gao L, Yuan Y, Liu ZT, Zhong YK, Zhi HJ (2015) Genetic analysis and identification of two soybean mosaic virus resistance genes in soybean [Glycine max (L.) Merr.]. Plant Breed 134(6):684–695
Li K, Yang QH, Zhi HH, Gai JY (2010) Identification and distribution of soybean mosaic virus strains in southern China. Plant Dis 94:351–357
Liu W, Li X, Zhou K, Pan X, Li Y, Lu T, Sheng X (2016) Mapping of QTLs controlling grain shape and populations construction derived from related residual heterozygous lines in rice. J Agric Sci 8:104–114
Ma G, Chen PY, Buss G, Tolin SA (2003) Genetic study of a lethal necrosis to soybean mosaic virus in PI 507389 soybean. J Hered 94:205–211
Ma Y, Li HC, Wang DG, Liu N, Zhi HJ (2010) Molecular mapping and marker assisted selection of soybean mosaic virus resistance gene RSC12 in soybean. Legume Genom Genet 1(8):1–6
Ma Y, Wang DG, Li HC, Zheng GJ, Yang YQ, Li HW, Zhi HJ (2011) Fine mapping of the RSC14Q locus for resistance to soybean mosaic virus in soybean. Euphytica 181:127–135
Maroof M, Tucker DM, Skoneczka JA, Bowman BC, Tripathy S, Tolin SA (2010) Fine mapping and candidate gene discovery of the soybean mosaic virus resistance gene, Rsv4. Plant Genome 3:14–22
Michelmore RW, Paran I, Kesseli R (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci 88:9828–9832
Mohan M, Nair S, Bhagwat A, Krishna T, Yano M, Bhatia C, Sasaki T (1997) Genome mapping, molecular markers and marker-assisted selection in crop plants. Mol Breed 3:87–103
Moon JK, Jeong SC, Van K, Maroof MS, Lee SH (2009) Marker-assisted identification of resistance genes to soybean mosaic virus in soybean lines. Euphytica 169:375–385
Pham AT, Harris DK, Buck J, Hoskins A, Serrano J, Abdel-Haleem H, Cregan P, Song QJ, Boerma HR, Li ZL (2015) Fine mapping and characterization of candidate genes that control resistance to Cercospora sojina K. Hara in two soybean germplasm accessions. PLoS ONE 10:e0126753
Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song QJ, Thelen JJ, Cheng JL, Xu D, Hellsten U, May GD, Yu Y, Sakurai T, Umezawa T, Bhattacharyya MK, Sandhu D, Valliyodan B, Lindquist E, Peto M, Grant D, Shu SQ, Goodstein D, Barry K, Futrell-Griggs M, Abernathy B, Du JC, Tian ZX, Zhu LC, Gill N, Joshi T, Libault M, Sethuraman A, Zhang XC, Shinozaki K, Nguyen HT, Wing RA, Cregan P, Specht J, Grimwood J, Rokhsar D, Stacey G, Shoemaker RC, Jackson SA (2010) Genome sequence of the palaeopolyploid soybean. Nature 463:178–183
Shirasawa K, Isobe S, Tabata S, Hirakawa H (2014) Kazusa Marker DataBase: a database for genomics, genetics, and molecular breeding in plants. Breed Sci 64:264–271
Song QJ, Marek L, Shoemaker R, Lark K, Concibido V, Delannay X, Specht JE, Cregan P (2004) A new integrated genetic linkage map of the soybean. Theor Appl Genet 109:122–128
Song QJ, Jia GF, Zhu YL, Grant D, Nelson RT, Hwang EY, Hyten DL, Cregan P (2010) Abundance of SSR motifs and development of candidate polymorphic SSR markers (BARCSOYSSR_1.0) in soybean. Crop Sci 50:1950–1960
Song J, Liu Z, Hong H, Ma Y, Tian L, Li X, Li Y-H, Guan R, Guo Y, Qiu L-J (2016) Identification and validation of loci governing seed coat color by combining association mapping and bulk segregation analysis in soybean. PLoS ONE 11(7):e0159064
Su CF, Lu WG, Zhao TJ, Gai JY (2010) Verification and fine-mapping of QTLs conferring days to flowering in soybean using residual heterozygous lines. Chin Sci Bull 55:499–508
Suh SJ, Bowman BC, Jeong N, Yang K, Kastl C, Tolin SA, Maroof M, Jeong SC (2011) The Rsv3 locus conferring resistance to soybean mosaic virus is associated with a cluster of coiled-coil nucleotide-binding leucine-rich repeat genes. Plant Genome 4:55–64
Tang S, Shao G, Wei X, Chen M, Sheng Z, Luo J, Jiao G, Xie L, Hu Tang S, Shao G, Wei X, Chen M, Sheng Z, Luo J, Jiao G, Xie L, Hu P (2013) QTL mapping of grain weight in rice and the validation of the QTL qTGW3.2. Gene 527:201–206
Tollenaere R, Hayward A, Dalton-Morgan J, Campbell E, Lee JR, Lorenc MT, Manoli S, Stiller J, Raman R, Raman H (2012) Identification and characterization of candidate Rlm4 blackleg resistance genes in Brassica napus using next-generation sequencing. Plant Biotechnol J 10:709–715
Tuyen D, Lal S, Xu DH (2010) Identification of a major QTL allele from wild soybean (Glycine soja Sieb. & Zucc.) for increasing alkaline salt tolerance in soybean. Theor Appl Genet 121:229–236
Van Ooijen J (2006) JoinMap 4. Software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen
Wang XQ, Gai JY, Pu ZQ (2003) Classification and distribution of strains of Soybean mosaic virus middle and lower Huang-Huai and Changjiang Valleys. Soybean Sci 22:102–107
Wang YJ, Dong Fang Y, Wang XQ, Yang YL, Yu DY, Gai JY, Wu XL, He CY, Zhang JS, Chen SY (2004) Mapping of five genes resistant to SMV strains in soybean. Acta Genet Sin 31:87–90
Wang YW, Zhi HJ, Guo DQ, Gai JY, Chen QS, Li K, Li HC (2005) Classification and distribution of strains of Soybean mosaic virus in northern china spring planting soybean region. Soybean Sci 24:263–268
Wang DG, Ma Y, Liu N, Yang ZL, Zheng GJ, Zhi HJ (2011a) Fine mapping of resistance to soybean mosaic virus strain SC4 in soybean based on genomic-SSR markers. Plant Breed 130:653–659
Wang DG, Ma Y, Yang YQ, Liu N, Li CY, Song YP, Zhi HJ (2011b) Fine mapping and analyses of RSC8 resistance candidate genes to soybean mosaic virus in soybean. Theor Appl Genet 122:555–565
Wang DG, Li HW, Zhi HJ, Tian Z, HuC, Hu GY, Huang ZP, Zhang L (2014) Identification of strains and screening of resistance resources to soybean mosaic virus in Anhui Province. Chin J Oil Crop Sci 36:374–379
Wang YF, Lu JJ, Chen S, Shu L, Palmer RG, Xing GN, Li Y, Yang SP, Yu DY, Zhao TJ, Gai JY (2014b) Exploration of presence/absence variation and corresponding polymorphic markers in soybean genome. J Integr Plant Biol 56:1009–1019
Xu R, Shi CE, Zhang LF (2004) Utilization of Qihuang 1 in soybean breeding in the Huanghuai region. J Plant Genet Resour 5:170–175
Yamanaka N, Watanabe S, Toda K, Hayashi M, Fuchigami H, Takahashi R, Harada K (2005) Fine mapping of the FT1 locus for soybean flowering time using a residual heterozygous line derived from a recombinant inbred line. Theor Appl Genet 110:634–639
Yamanaka N, Fuentes F, Gilli J, Watanabe S, Harada K, Ban T, Abdelnoor R, Nepomuceno A, Homma Y (2006) Identification of quantitative trait loci for resistance against soybean sudden death syndrome caused by Fusarium tucumaniae. Pesqui Agropecu Bras 41:1385–1391
Yan H, Wang H, Cheng H, Hu Z, Chu S, Zhang G, Yu D (2015) Detection and fine-mapping of SC7 resistance genes via linkage and association analysis in soybean. J Integr Plant Biol 57:722–729
Yang QH, Gai JY (2010) Identification, inheritance and gene mapping of resistance to a virulent soybean mosaic virus strain SC15 in soybean. Plant Breed 130:128–132
Yang YQ, Zheng GJ, Han L, Wang DG, Yang XF, Yuan Y, Huang SH, Zhi HJ (2013) Genetic analysis and mapping of genes for resistance to multiple strains of Soybean mosaic virus in a single resistant soybean accession PI 96983. Theor Appl Genet 126:1783–1791
You FM, Huo N, Gu YQ, Luo MC, Ma Y, Hane D, Lazo GR, Dvorak J, Anderson OD (2008) BatchPrimer3: a high through put web application for PCR and sequencing primer design. BMC Bioinform 9:1
Yu YG, Saghai MMA, Buss GR, Maughan PJ, Tolin SA (1994) RFLP and microsatellite mapping of a gene for soybean mosaic virus resistance. Phytopathology 84:60–64
Zhan Y, Zhi HJ, Yu DY, Gai JY (2006) Identification and distribution of SMV strains in Huang-Huai Valleys. Sci Agric Sin 39:2009–2015
Zheng GJ, Yang YQ, Ma Y, Yang XF, Chen SY, Ren R, Wang DG, Yang ZL, Zhi HJ (2014) Fine mapping and candidate gene analysis of resistance gene RSC3Q to soybean mosaic virus in Qihuang No. 1. J Integr Agric 13:2608–2615
Acknowledgements
This work was financially supported through grants from the China National Key R & D Program for crop Breeding (2016YFD0100304, 2017YFD0101500), the Natural Science Foundation of China (31671718, 31571695), the MOE 111 Project (B08025), the MOE Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT_17R55), the MOA CARS-04 program, the Jiangsu Higher Education PAPD Program, the Fundamental Research Funds for the Central Universities (Y0201600115) and the Jiangsu JCIC-MCP. The funders had no role in work design, data collection and analysis, or decision and preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared that no competing or conflicts of interest exist.
Ethical standards
The experiments were performed in compliance with the current laws of China.
Additional information
Communicated by Henry T. Nguyen.
Rights and permissions
About this article
Cite this article
Karthikeyan, A., Li, K., Li, C. et al. Fine-mapping and identifying candidate genes conferring resistance to Soybean mosaic virus strain SC20 in soybean. Theor Appl Genet 131, 461–476 (2018). https://doi.org/10.1007/s00122-017-3014-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-017-3014-1