Abstract
Key message
The newly discovered determinate plant growth habit of Brassica napus is a potential trait that might contribute to the genetic improvement of rapeseed.
Abstract
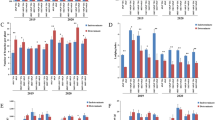
Brassica napus is an important species of rapeseed and has an indeterminate growth habit. However, a determinate inflorescence strain (4769) has been discovered among doubled haploid (DH) lines obtained from a spring B. napus × winter B. napus cross. We assessed the effect of the determinate growth habit on agronomic traits. The results showed that determinacy is beneficial for reducing plant height and flowering time, advancing maturity and maintaining productivity. We also investigated the inheritance of determinacy. A genetic analysis revealed that the phenotype of the determinate trait is controlled by one recessive gene, Bnsdt1. Mapping of the Bnsdt1 gene was subsequently conducted in BC1 and BC3 populations derived from combination 2014 × 4769. The results showed that the Bnsdt1 gene could be delimited to a region of approximately 220 kb, between 16,627 and 16,847 kb on A10. Within the target region, whole-genome re-sequencing identified two candidate regions (16,628–16,641 and 16,739–16,794 kb) of approximately 68 kb. A Blast analysis of the two candidate intervals found that BnaA10g26300D/GSBRNA2T00136426001 (BnTFL1) is homologous to the TFL1 gene of A. thaliana. Subsequently, quantitative reverse transcription (qRT)-PCR revealed that BnTFL1 was specifically expressed in the shoot apex. Collectively, the results of expression analysis provide preliminary evidence that BnTFL1 is a candidate gene for the inflorescence trait in 4769.









Similar content being viewed by others
References
Alvarez J, Guli CL, Yu XH, Smyth DR (1992) terminal flower: a gene affecting inflorescence development in Arabidopsis thaliana. Plant J 2(1):103–116
Amasino R (2010) Seasonal and developmental timing of flowering. Plant J 61(6):1001–1013
Amaya I, Ratcliffe OJ, Bradley DJ (1999) Expression of CENTRORADIALIS (CEN) and CEN-like genes in tobacco reveals a conserved mechanism controlling phase change in diverse species. Plant Cell 11(8):1405–1417
Banga SS (2007) Genetic manipulations for oilseeds improvement—conventional. In: Hedge DM (ed) Changing global vegetable oils scenario: issues and challenges before India. ISOR, Hyderabad, pp 17–34
Banga SS, Kaur N (2009) An alternate procedure for resynthesis of Brassica juncea. In: Proceedings of 16th Australian research assembly on Brassicas, Ballarat, vol 106, pp 1–4
Bansal P, Kaur P, Banga SK, Banga SS (2009) Augmenting genetic diversity in Brassica juncea through its resynthesis using purposely selected diploid progenitors. Int J Plant Breed 3:41–45
Bradley D, Carpenter R, Copsey L, Vincent C (1996) Control of inflorescence architecture in Antirrhinum. Nature 376:791–797
Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E (1997) Inflorescence commitment and architecture in Arabidopsis. Science 275(5296):80–83
Bratzel F, Turck F (2015) Molecular memories in the regulation of seasonal flowering: from competence to cessation. Genome Biol 16(1):192
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Gupta M, Atri C, Banga SS (2014) Cytogenetic stability and genome size variations in newly developed derived Brassica juncea allopolyploid lines. Jour Oilseed Brassica 5:118–127
Hanano S, Goto K (2011) Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell 23:3172–3184
Hanzawa Y, Money T, Bradley D (2005) A single amino acid converts a repressor to an activator of flowering. Proc Natl Acad Sci USA 102:7748–7753
He J, Ke L, Hong D, Xie Y, Wang G, Liu P, Yang G (2008) Fine mapping of a recessive genic male sterility gene (Bnms3) in rapeseed (Brassica napus) with AFLP- and Arabidopsis-derived PCR markers. Theor Appl Genet 117(1):11–18
Huijser P, Schmid M (2011) The control of developmental phase transitions in plants. Development 138(19):4117–4129
Kaur H, Banga SS (2015) Discovery and mapping of Brassica juncea Sdt1 gene associated with determinate plant growth habit. Theor Appl Genet 128(2):235–245
Kaur H, Gupta S, Kumar N, Akhatar J, Banga SS (2014) Progression of molecular and phenotypic diversification in resynthesized Brassica juncea (L) gene pool with determinate inflorescence. Euphytica 199(3):325–338
Laucou V, Haurogne K, Ellis N, Rameau C (1998) Genetic mapping in pea. 1. RAPD-based genetic linkage map of Pisum sativum. Theor Appl Genet 97(5–6):905–915
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760
Li Q, Wan JM (2005) SSRHunter: development of a local searching software for SSR sites. Hereditas (Beijing) 27:808–810
Liu HL (2000) Genetics and breeding in rapeseed. Chinese Agricultural University Press, Beijing, p 49
Liu RH, Meng JL (2003) MapDraw: a microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Genetics 25(3):317–321
Lowe AJ, Jones AE, Raybould AF, Trick M, Moule CJ, Edwards KJ (2002) Transferability and genome specificity of a new set of microsatellite primers among Brassica species of the U triangle. Mol Ecol Notes 2:7–11
Lu H, Lin T, Klein J, Wang S, Qi JJ, Zhou Q, Sun JJ, Zhang ZH, Weng YQ, Huang SW (2014) QTL-seq identifies an early flowering QTL located near Flowering Locus T in cucumber. Theor Appl Genet 127:1491–1499. doi:10.1007/s00122-014-2313-z
McKenna A, Hanna M, Banks EM, Sivachenko A, Cibulskis K, Kernytsky A, DePristo MA (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20(9):1297–1303
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci 88:9828–9832
Norusis M (2008) SPSS 16.0 statistical procedures companion. Prentice Hall, New Jersey
Panjabi P, Jagannath A, Bisht NC, Padmaja KL, Sharma S, Gupta V, Pental D (2008) Comparative mapping of Brassica juncea and Arabidopsis thaliana using Intron Polymorphism (IP) markers: homoeologous relationships, diversification and evolution of the A, B and C Brassica genomes. BMC Genom 9(1):113
Pnueli L, Gutfiger T, Hareven D, Ben-Naim O, Ron N, Adir N, Lifschitz E (2001) Tomato SP–interacting proteins define a conserved signaling system that regulates shoot architecture and flowering. Plant Cell 13:2687–2702
Quijada PA, Udall JA, Lambert B, Osborn TC (2006) Quantitative trait analysis of seed yield and other complex traits in hybrid spring rapeseed (Brassica napus L.): 1. Identification of genomic regions from winter germplasm. Theor Appl Genet 113:549–561
Shannon S, Meeks-Wagner DR (1991) A mutation in the Arabidopsis TFL1 gene affects inflorescence meristem development. Plant Cell 3:877–892
Srikanth A, Schmid M (2011) Regulation of flowering time: all roads lead to Rome. Cell Mol Life Sci 68(12):2013–2037
Tahery Y, Abhul-Hamid H, Tahery E (2011) Terminal Flower 1 (TFL1) homolog genes in dicot plants. World Appl Sci J 12(545):551
Torti S, Fornara F, Vincent C, Andrés F, Nordström K, Göbel U, Coupland G (2012) Analysis of the Arabidopsis shoot meristem transcriptome during floral transition identifies distinct regulatory patterns and a leucine-rich repeat protein that promotes flowering. Plant Cell 24(2):444–462
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth C, Remm M, Rozen SG (2012) Primer3-new capabilities and interfaces. Nucleic Acids Res 40(15):115
Van Ooijen JW (2006) JoinMap 4: Software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen
Vos P, Hogers R, Bleeker M, Reijans M, Van de Lee T, Hornes M, Freijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Xiao L, Zhao Z, Du DZ, Yao YM, Xu L, Tang GY (2012) Genetic characterization and fine mapping of a yellow-seeded gene in Dahuang (a Brassica rapa landrace). Theor Appl Genet 124(5):903–909
Yi B, Chen YN, Lei SL, Tu JX, Fu TD (2006) Fine mapping of the recessive genic male-sterile gene (Bnms1) in Brassica napus L. Theor Appl Genet 113:643–650
Zhang HY, Miao HM, Li C, Wei LB, Duan YH, Ma Q, Chang SX (2016) Ultra-dense SNP genetic map construction and identification of SiDt gene controlling the determinate growth habit in Sesamum indicum L. Sci Rep. doi:10.1038/s41598-017-02405-9
Zhao Z, Xiao L, Xu L, Xing X, Tang G, Du D (2017) Fine mapping the BjPl1 gene for purple leaf color in B2 of Brassica juncea L. through comparative mapping and whole-genome re-sequencing. Euphytica 213(4):1–11
Acknowledgements
This research was financially supported by the National System of Technology of the Rapeseed Industry (NYCYTX-00516), the National Key Research and Development Plan of China (2016YFD0100202) and Key Laboratory of Spring Rape Genetic Improvement of Qinghai Province (2017-ZJ-Y09).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
The authors declare that the study complies with the current laws of China.
Additional information
Communicated by Maria Laura Federico.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, K., Yao, Y., Xiao, L. et al. Fine mapping of the Brassica napus Bnsdt1 gene associated with determinate growth habit. Theor Appl Genet 131, 193–208 (2018). https://doi.org/10.1007/s00122-017-2996-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-017-2996-z