Abstract
Key message
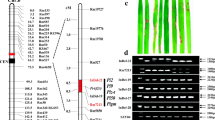
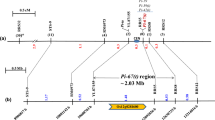
A novel R gene was mapped to a locus on chromosome 11 from 30.42 to 30.85 Mb, which was proven to be efficient in the improvement of rice blast resistance.
Abstract
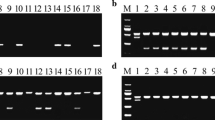
Rice blast is a devastating fungal disease worldwide. The use of blast resistance (R) genes is the most important approach to control the disease in rice breeding. In the present study, we finely mapped a novel resistance gene Pi65(t), conferring a broad-spectrum resistance to the fungus Magnaporthe oryzae, using bulked segregant analysis in combination with next-generation sequencing technology. Segregation in a doubled haploid (DH) population and a BC1F2 population suggested that resistance to blast in Gangyu129 was likely conferred by a single dominant gene, designated Pi65(t); it was located on chromosome 11 from 30.20 to 31.20 Mb using next-generation sequencing. After screening recombinants with newly developed molecular markers, the region was narrowed down to 0.43 Mb, flanked by SNP-2 and SNP-8 at the physical location from 30.42 to 30.85 Mb based on the Nipponbare reference database in build 5. Using the software QTL IciMapping, Pi65(t) was further mapped to a locus between InDel-1 and SNP-4 with genetic distances of 0.11 and 0.98 cM, respectively. Within this region, 4 predicted R genes were found with nucleotide binding site and leucine-rich repeat (NBS-LRR) domains. We developed molecular markers to genotype 305 DH lines and found that InDel-1 was closely linked with Pi65(t). Using InDel-1, a new rice variety Chuangxin1 containing Pi65(t) was developed, and it is highly resistant to rice blast and produces a high yield in Liaoning province of China. This indicated that Pi65(t) could play a key role in the improvement of rice blast resistance.



Similar content being viewed by others
References
Abe A, Kosugi S, Yoshida K et al (2012) Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol 30:174–178
Ashikawa I, Hayashi N, Yamane H, Kanamori H, Wu JZ, Matsumoto T, One K, Yano M (2008) Two adjacent nucleotide-binding site-leucine-rich repeat class genes are required to confer Pikm-specific rice blast resistance. Genetics 180:2267–2276
Boyd LA, Ridout C, O’Sullivan DM, Leach JE, Leung H (2013) Plant–pathogen interactions: disease resistance in modern agriculture. Trends Genet 29:233–240
Chauhan RS, Farman ML, Zhang HB, Leong SA (2002) Genetic and physical mapping of a rice blast resistance locus, Pi-CO39(t), that corresponds to the avirulence gene AVR1-CO39 of Magnaporthe grisea. Mol Genet Genom 267:603–612
Chen DH, Zeigler RS, Leung H, Nelson RJ (1995) Population structure of Pyricularia grisea at two screening sites in the Philippines. Phytopathology 85:1011–1020
Chen W, Yao JB, Chu L, Yuan ZW, Li Y, Zhang YS (2015) Genetic mapping of the nulliplex-branch gene (gb_nb1) in cotton using next-generation sequencing. Theor Appl Genet 128:539–547
Cheng LR, Yang AG, Jiang CH, Ren M, Zhang Y, Feng QF, Wang SM, Guan YS, Luo CG (2015) Quantitative trait loci mapping for plant height in tobacco using linkage and association mapping methods. Crop Sci 55:641–647
Das S, Upadhyaya H, Bajaj D et al (2015) Deploying QTL-seq for rapid delineation of a potential candidate gene underlying major trait-associated QTL in chickpea. DNA Res. doi:10.1093/dnares/dsv004
Dean RA, Talbot NJ, Ebbole DJ et al (2005) The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434:980–986
Fekih R, Takagi H, Tamiru M et al (2013) MutMap+: genetic mapping and mutant identification without crossing in rice. PLoS ONE 8:e68529
Fukuoka S, Saka N, Koga H et al (2009) Loss of function of a proline-containing protein confers durable disease resistance in rice. Science 325:998–1001
Hayashi K, Yoshida H, Ashikawa I (2006) Development of PCR-based allele-specific and InDel marker sets for nine rice blast resistance genes. Theor Appl Genet 113:251–260
Hittalmani S, Parco A, Mew TV, Zeigler RS, Huang N (2000) Fine mapping and DNA marker-assisted pyramiding of the three major genes for blast resistance in rice. Theor Appl Genet 100:1121–1128
Huang HM, Huang L, Feng GP et al (2011) Molecular mapping of the new blast resistance genes Pi47 and Pi48 in the durably resistant local rice cultivar Xiangzi 3150. Phytopathology 101:620–626
Hulbert SH, Webb CA, Smith SM, Sun Q (2001) Resistance gene complexes: evolution and utilization. Annu Rev Phytopathol 39:285–312
Jeon JS, Chen D, Yi GH, Wang G, Ronald P (2003) Genetic and physical mapping of Pi5 (t), a locus associated with broad-spectrum resistance to rice blast. Mol Genet Genom 269:280–289
Jeung JU, Kim BR, Cho YC, Han SS, Moon HP, Lee YT, Jena KK (2007) A novel gene, Pi40(t), linked to the DNA markers derived from NBS-LRR motifs confers broad spectrum of blast resistance in rice. Theor Appl Genet 115:1163–1177
Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B (2000) Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J 19:4004–4014
Jia YL, Wang ZH, Singh P (2002) Development of dominant rice blast Pi-ta resistance gene markers. Crop Sci 42:2145–2149
Kent WJ (2002) BLAT—the BLAST-like alignment tool. Genome Res 12:656–664
Kosambi DD (1943) The estimation of map distance from recombination values. Ann Eugen 12:172–175
Li HH, Ribaut JM, Li ZL, Wang JK (2008) Inclusive composite interval mapping (ICIM) for digenic epistasis of quantitative traits in biparental populations. Theor Appl Genet 116:243–260
Liu XQ, Wang L, Chen S, Lin F, Pan QH (2005) Genetic and physical mapping of Pi36 (t), a novel rice blast resistance gene located on rice chromosome 8. Mol Genet Genomics 274:394–401
Liu JL, Wang XJ, Mitchell T, Hu YJ, Liu XL, Dai LY, Wang GL (2010) Recent progress and understanding of the molecular mechanisms of the rice–Magnaporthe oryzae interaction. Mol Plant Pathol 11:419–427
Liu WD, Liu JL, Ning YS, Ding B, Wang XL, Wang ZL, Wang GL (2013a) Recent progress in understanding PAMP- and effector-triggered immunity against the rice blast Fungus Magnaporthe oryzae. Mol Plant 6:605–620
Liu Y, Liu B, Zhu XY, Yang JY, Bordeos A, Wang GL, Leach JE, Leung H (2013b) Fine-mapping and molecular marker development for Pi56(t), a NBS-LRR gene conferring broad-spectrum resistance to Magnaporthe oryzae in rice. Theor Appl Genet 126:985–998
Liu Y, Qi XS, Young ND, Olsen KM, Caicedo AL, Jia YL (2015) Characterization of resistance genes to rice blast fungus Magnaporthe oryzae in a “Green Revolution” rice variety. Mol Breeding 35:1–8
Livaja M, Wang Y, Wieckhorst S, Haseneyer Grit et al (2013) BSTA: a targeted approach combines bulked segregant analysis with next-generation sequencing and de novo transcriptome assembly for SNP discovery in sunflower. BMC Genom 14:628
Lu HF, Lin T, Klein J, Wang SH, Qi JJ, Zhou Q, Sun JJ, Zhang ZH, Weng YQ, Huang SW (2014) QTL-seq identifies an early flowering QTL located near flowering locus T in cucumber. Theor Appl Genet 127:1491–1499
Ma J, Jia M, Jia Y (2014) Characterization of rice blast resistance gene Pi61 (t) in rice germplasm. Plant Dis 98:1200–1204
Michelmore RW, Paran I, Kesseli R (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Am Sci USA 88:9828–9832
Ou SH (1985) Rice diseases, 2nd edn. Commonwealth Mycological Institute, Kew
Pan QH, Wang L, Ikehashi H, Tanisaka T (1996) Identification of a new blast resistance gene in the indica rice cultivar Kasalath using Japanese differential cultivars and isozyme markers. Phytopathology 86:1071–1075
Qu SH, Liu GF, Zhou B, Bellizzi M, Zeng L, Dai LY, Han B, Wang GL (2006) The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site–leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 172:1901–1914
Salvi S, Tuberosa R (2005) To clone or not to clone plant QTLs: present and future challenges trends in plant. Science 10:297–304
Sharma T, Rai A, Gupta S, Singh N (2010) Broad-spectrum blast resistance gene Pi-kh cloned from rice line tetep designated as Pi54. J Plant Biochem Biot 19:87–89
Sun XW, Liu DY, Zhang XF et al (2013) SLAF-seq: an efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS ONE 8:e58700
Takagi H (2013) QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J 74:174–183
Takagi H, Uemura A, Yaegashi H et al (2013) MutMap-Gap: whole-genome resequencing of mutant F2 progeny bulk combined with de novo assembly of gap regions identifies the rice blast resistance gene Pii. New Phytol 200:276–283
Takagi H, Tamiru M, Abe A et al (2015) MutMap accelerates breeding of a salt-tolerant rice cultivar. Nat Biotechnol 33:445–449
Trick M, Adamski NM, Mugford SG, Jiang CC, Febrer M, Uauy C (2012) Combining SNP discovery from next-generation sequencing data with bulked segregant analysis (BSA) to fine-map genes in polyploid wheat. BMC Plant Biol 12:14
Wang SW, Zheng WJ, Zhao JM, Wei SH, Wang Y, Zhao BH, Liu ZH (2013) Identification and analysis of Magnaporthe oryzae avirulence genes in Liaoning Province. Scientia Agricultura Sinica 47:462–472 (In Chinese)
Wang Y, Zhao JM, Zhang LX, Wang P, Wang SW, Wang H, Wang XX, Liu ZH, Zheng WJ (2015) Analysis of the diversity and function of the alleles of the rice blast resistance genes Piz-t, Pita and Pik in 24 rice cultivars. J Integr Agr. doi:10.1016/S2095-3119(15)61207-2
Xia C, Chen LL, Rong TZ, Li R, Xiang Y, Wang P, Liu CH, Dong XQ, Liu B, Zhao D, Wei RJ, Lan H (2014) Identification of a new maize inflorescence meristem mutant and association analysis using SLAF-seq method. Euphytica 202:35–44
Xu X, Hayashi N, Wang CT, Fukuoka S, Kawasaki S, Takatsuji H, Jiang CJ (2014) Rice blast resistance gene Pikahei-1 (t), a member of a resistance gene cluster on chromosome 4, encodes a nucleotide-binding site and leucine-rich repeat protein. Mol Breed 34:691–700
Yang ZM, Huang DQ, Tang WQ, Zheng Y, Liang KJ, Cutler AJ, Wu WR (2013) Mapping of quantitative trait loci underlying cold tolerance in rice seedlings via high-throughput sequencing of pooled extremes. PLoS ONE 8:e68433
Zuo WL, Chao Q, Zhang N et al (2014) A maize wall-associated kinase confers quantitative resistance to head smut. Nat Genet 2015 47:151–157
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No.31571993 and 31301636), and Natural science foundation of Liaoning Province in China (2014027027).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest in regard to this manuscript.
Ethical standards
We declare that these experiments comply with the ethical standards in China, where they were performed.
Additional information
Communicated by M. Thomson.
W. Zheng and Y. Wang contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zheng, W., Wang, Y., Wang, L. et al. Genetic mapping and molecular marker development for Pi65(t), a novel broad-spectrum resistance gene to rice blast using next-generation sequencing. Theor Appl Genet 129, 1035–1044 (2016). https://doi.org/10.1007/s00122-016-2681-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-016-2681-7