Abstract
Key message
Genomic selection empirically appeared valuable for reciprocal recurrent selection in oil palm as it could account for family effects and Mendelian sampling terms, despite small populations and low marker density.
Abstract
Genomic selection (GS) can increase the genetic gain in plants. In perennial crops, this is expected mainly through shortened breeding cycles and increased selection intensity, which requires sufficient GS accuracy in selection candidates, despite often small training populations. Our objective was to obtain the first empirical estimate of GS accuracy in oil palm (Elaeis guineensis), the major world oil crop. We used two parental populations involved in conventional reciprocal recurrent selection (Deli and Group B) with 131 individuals each, genotyped with 265 SSR. We estimated within-population GS accuracies when predicting breeding values of non-progeny-tested individuals for eight yield traits. We used three methods to sample training sets and five statistical methods to estimate genomic breeding values. The results showed that GS could account for family effects and Mendelian sampling terms in Group B but only for family effects in Deli. Presumably, this difference between populations originated from their contrasting breeding history. The GS accuracy ranged from −0.41 to 0.94 and was positively correlated with the relationship between training and test sets. Training sets optimized with the so-called CDmean criterion gave the highest accuracies, ranging from 0.49 (pulp to fruit ratio in Group B) to 0.94 (fruit weight in Group B). The statistical methods did not affect the accuracy. Finally, Group B could be preselected for progeny tests by applying GS to key yield traits, therefore increasing the selection intensity. Our results should be valuable for breeding programs with small populations, long breeding cycles, or reduced effective size.






Similar content being viewed by others
References
Billotte N, Marseillac N, Risterucci AM et al (2005) Microsatellite-based high density linkage map in oil palm (Elaeis guineensis Jacq.). Theor Appl Genet 110:754–765
Browning SR, Browning BL (2007) Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet 81:1084–1097
Butler DG, Cullis BR, Gilmour AR, Gogel BJ (2009) Mixed models for S language environments: ASReml-R reference manual (Version 3). Queensland Department of Primary Industries and Fisheries
Cochard B (2008) Etude de la diversité génétique et du déséquilibre de liaison au sein de populations améliorées de palmier à huile (Elaeis guineensis Jacq.). Montpellier SupAgro, Montpellier, pp 97–175
Cochard B, Adon B, Rekima S et al (2009) Geographic and genetic structure of African oil palm diversity suggests new approaches to breeding. Tree Genet Genomes 5:493–504
Corley RHV (2009) How much palm oil do we need? Environ Sci Policy 12:134–139
Corley RHV, Tinker PB (2003) Selection and breeding. The oil palm, 4th edn. Blackwell Science Ltd Blackwell Publishing, Oxford, pp 133–199
Cros D, Sánchez L, Cochard B et al (2014) Estimation of genealogical coancestry in plant species using a pedigree reconstruction algorithm and application to an oil palm breeding population. Theor Appl Genet 127:981–994
Daetwyler HD, Villanueva B, Bijma P, Woolliams JA (2007) Inbreeding in genome-wide selection. J Anim Breed Genet 124:369–376
Daetwyler HD, Calus MPL, Pong-Wong R, de los Campos G, Hickey JM (2013) Genomic prediction in animals and plants: simulation of data, validation, reporting, and benchmarking. Genetics 193:347–365
de los Campos G, Pérez P, Vazquez A, Crossa J (2013) Genome-enabled prediction using the BLR (Bayesian linear regression) R-Package. In: Gondro C, van der Werf J, Hayes B (eds) Genome-wide association studies and genomic prediction. Humana Press, New York, pp 299–320
de los Campos G, Naya H, Gianola D et al (2009) Predicting quantitative traits with regression models for dense molecular markers and pedigree. Genetics 182:375–385
Dussert S, Guerin C, Andersson M et al (2013) Comparative transcriptome analysis of three oil palm fruit and seed tissues that differ in oil content and fatty acid composition. Plant Physiol 162:1337–1358
Eding H, Meuwissen THE (2001) Marker-based estimates of between and within population kinships for the conservation of genetic diversity. J Anim Breed Genet 118:141–159
Gao H, Lund MS, Zhang Y, Su G (2013) Accuracy of genomic prediction using different models and response variables in the Nordic Red cattle population. J Anim Breed Genet 130:333–340
Garrick D, Taylor J, Fernando R (2009) Deregressing estimated breeding values and weighting information for genomic regression analyses. Genet Sel Evol 41:55
Gascon JP, de Berchoux C (1964) Caractéristique de la production d’Elaeis guineensis (Jacq.) de diverses origines et de leurs croisements. Application à la sélection du palmier à huile. Oleagineux 19:75–84
Grattapaglia D (2014) Breeding forest trees by genomic selection: current progress and the way forward. In: Tuberosa R, Graner A, Frison E (eds) Genomics of plant genetic resources. Springer, Netherlands, pp 651–682
Habier D, Fernando RL, Dekkers JCM (2007) The impact of genetic relationship information on genome-assisted breeding values. Genetics 177:2389–2397
Habier D, Tetens J, Seefried F-R, Lichtner P, Thaller G (2010) The impact of genetic relationship information on genomic breeding values in German Holstein cattle. Genet Sel Evol 42:5
Habier D, Fernando R, Kizilkaya K, Garrick D (2011) Extension of the bayesian alphabet for genomic selection. BMC Bioinform 12:186
Henderson C (1975) Best linear unbiased estimation and prediction under a selection model. Biometrics 31:423–447
Heslot N, Yang H-P, Sorrells ME, Jannink J-L (2012) Genomic selection in plant breeding: a comparison of models. Crop Sci 52:146–160
Isik F (2014) Genomic selection in forest tree breeding: the concept and an outlook to the future. New Forest 45:379–401
Jannink J-L, Lorenz AJ, Iwata H (2010) Genomic selection in plant breeding: from theory to practice. Brief Funct Genom 9:166–177
Kumar S, Chagné D, Bink MCAM, Volz RK, Whitworth C, Carlisle C (2012) Genomic selection for fruit quality traits in apple (Malus domestica Borkh.). PLoS ONE 7:e36674
Li CC, Weeks DE, Chakravarti A (1993) Similarity of DNA fingerprints due to chance and relatedness. Hum Hered 43:45–52
Lorenz AJ, Chao S, Asoro FG et al (2011) Genomic selection in plant breeding: knowledge and prospects. In: Sparks DL (ed) Advances in agronomy. Academic Press, San Diego, pp 77–123
Lynch M (1988) Estimation of relatedness by DNA fingerprinting. Mol Biol Evol 5:584–599
Meuwissen THE, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829
Ostersen T, Christensen O, Henryon M, Nielsen B, Su G, Madsen P (2011) Deregressed EBV as the response variable yield more reliable genomic predictions than traditional EBV in pure-bred pigs. Genet Sel Evol 43:38
Park T, Casella G (2008) The Bayesian LASSO. J Am Stat Assoc 103:681–686
Pérez P, de los Campos G, Crossa J, Gianola D (2010) Genomic-enabled prediction based on molecular markers and pedigree using the Bayesian linear regression package in R. Plant Genome 3:106–116
Purba AR, Flori A, Baudouin L, Hamon S (2001) Prediction of oil palm (Elaeis guineensis, Jacq.) agronomic performances using the best linear unbiased predictor (BLUP). Theor Appl Genet 102:787–792
R Core Team (2013) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org
Resende MDV, Resende MFR, Sansaloni CP et al (2012) Genomic selection for growth and wood quality in Eucalyptus: capturing the missing heritability and accelerating breeding for complex traits in forest trees. New Phytol 194:116–128
Rincent R, Laloe D, Nicolas S et al (2012) Maximizing the reliability of genomic selection by optimizing the calibration set of reference individuals: comparison of methods in two diverse groups of maize inbreds (Zea mays L.). Genetics 192:715–728
Saatchi M, McClure M, McKay S et al (2011) Accuracies of genomic breeding values in American Angus beef cattle using K-means clustering for cross-validation. Genet Sel Evol 43:40
Singh R, Ong-Abdullah M, Low E-TL et al (2013) Oil palm genome sequence reveals divergence of interfertile species in Old and New worlds. Nat Adv. doi:10.1038/nature12309
Solberg TR, Sonesson AK, Woolliams JA, Meuwissen THE (2008) Genomic selection using different marker types and densities. J Anim Sci 86:2447–2454
Stuber CW, Cockerham CC (1966) Gene effects and variances in hybrid populations. Genetics 54:1279–1286
Tee S-S, Tan Y-C, Abdullah F, Ong-Abdullah M, Ho C-L (2013) Transcriptome of oil palm (Elaeis guineensis Jacq.) roots treated with Ganoderma boninense. Tree Genet Genom 9:377–386
Thomsen H, Reinsch N, Xu N et al (2001) Comparison of estimated breeding values, daughter yield deviations and de-regressed proofs within a whole genome scan for QTL. J Anim Breed Genet 118:357–370
Tranbarger TJ, Dussert S, Joët T et al (2011) Regulatory mechanisms underlying oil palm fruit mesocarp maturation, ripening, and functional specialization in lipid and carotenoid metabolism. Plant Physiol 156:564–584
Tranbarger T, Kluabmongkol W, Sangsrakru D et al (2012) SSR markers in transcripts of genes linked to post-transcriptional and transcriptional regulatory functions during vegetative and reproductive development of Elaeis guineensis. BMC Plant Biol 12:1
USDA (2013) Oilseeds: world market and trade. Foreign Agricultural Service, Circular Series, July 2013 http://www.fas.usda.gov/oilseeds_arc.asp
Waples RS, Do CHI (2008) LDNE: a program for estimating effective population size from data on linkage disequilibrium. Mol Ecol Resour 8:753–756
Wong CK, Bernardo R (2008) Genomewide selection in oil palm: increasing selection gain per unit time and cost with small populations. Theor Appl Genet 116:815–824
Zapata-Valenzuela J, Isik F, Maltecca C et al (2012) SNP markers trace familial linkages in a cloned population of Pinus taeda: prospects for genomic selection. Tree Genet Genom 8:1307–1318
Acknowledgments
We acknowledge SOCFINDO (Indonesia) and CRAPP (Benin) for planning and carrying out the field trials with CIRAD (France) and authorizing use of the phenotypic data for this study. This research was partly funded by a Grant from PalmElit SAS. We thank P. Sampers, C. Carrasco-Lacombe, A. Manez, and S. Tisné for their help in genotyping, L. Dedieu for reviewing the manuscript as well as two anonymous reviewers and C.C. Schön for their helpful comments.
Conflict of interest
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Chris Carolin Schön.
Electronic supplementary material
Below is the link to the electronic supplementary material.
122_2014_2439_MOESM2_ESM.tif
Figure S2 Pedigree of the 131 Group B individuals used in the study (in colors, with dark blue for La Mé population, green for Yangambi, yellow for Nigeria and light blue for La Mé × Yangambi and La Mé × Sibiti) (TIFF 9019 kb)
122_2014_2439_MOESM3_ESM.tif
Figure S3 Correlations between accuracies of the five statistical methods used to obtain GEBV (BayesCπ, BayesDπ, Bayesian Lasso regression [BL], Bayesian Ridge regression [BRR] and GBLUP). Accuracy was calculated as the correlation between GEBV and EBV in the test set. One dot is the value obtained for one test set, for a combination of population (Deli or Group B), trait (eight studied traits), definition of training set (three methods) and replicate (one for CDmean and five for clustering and Within-Family). The diagonal line is the y = x line (TIFF 22 kb)
122_2014_2439_MOESM4_ESM.tif
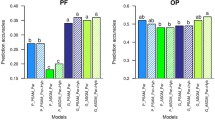
Figure S4 Diagram of interactions between trait and population on the GBLUP accuracy. Values are means over 11 accuracy estimates (five for clustering, five for Within-Family and one for CDmean). Values with the same letters are not significantly different at P = 0.001 (TIFF 33 kb)
122_2014_2439_MOESM5_ESM.tif
Figure S5 Narrow sense heritability (h 2) for eight yield traits (ABW: average bunch weight, BN: bunch number, FW: fruit weight, NF: number of fruits per bunch, F/B: fruits to bunch ratio, P/F: pulp to fruit ratio, O/P: oil to pulp ratio and K/F: kernel to fruit ratio) estimated from progeny tests between Deli and Group B (TIFF 18 kb)
122_2014_2439_MOESM6_ESM.tif
Figure S6 Accuracy of GBLUP versus bias in test sets for the GBLUP method. One dot is the value obtained for one test set, for a combination of population (Deli or Group B), trait (eight studied traits), definition of training set (CDmean, K-means clustering and Within-Family) and replicate (one for CDmean and five for clustering and Within-Family). The dashed grey line indicates unbiased GEBV (TIFF 13 kb)
Appendix: Estimation of parental breeding values
Appendix: Estimation of parental breeding values
The mating design of the progeny tests consisted of 445 Deli × Group B crosses made according to an incomplete factorial design. The crosses were evaluated in 26 trials planted between 1995 and 2000. The experimental designs of the trials were RCBD with five or six blocks and balanced lattices of rank four or five. The bunch production was measured on 30,872 palms and bunch quality on 21,525 palms. Eight traits were studied. The bunch number (BN) and average bunch weight (ABW) were measured every ten days on palms from ages 6 to 11. The annual cumulative BN and mean annual ABW were used in analysis. The median number of progenies with bunch production data was 169 per Deli parent (ranging from 25 to 743) and 141 (23–859) per Group B parent. The fruit-to-bunch (F/B), pulp-to-fruit (P/F), kernel-to-fruit (K/F), and oil-to-pulp (O/P) ratios, the number of fruits per bunch (NF), and the average fruit weight (FW) were measured on two bunches at ages five and six on a sample of at least 24 palms per cross. The median number of bunches analyzed was 327 per Deli parent (ranging from 69 to 1,358) and 309 per Group B parent (73–1,149).
EBV were computed as traditional pedigree-based BLUP (T-BLUP) predictors of the random effects a A and a B , using a mixed model of the form:
where y is the vector of data records for the trait being analyzed, β the vector of fixed effects (general mean, trial and block within trial), a Deli and a B vectors of general combining ability of Deli ~N(0, 0.5A Deli \(\sigma_{\rm{Deli}}^{2}\)) and Group B individuals ~N(0, 0.5A B \(\sigma_{B}^{2}\)), respectively, b the vector of the incomplete block within block and trial effects ~N(0, I \(\sigma_{b}^{2}\)), c the vector of specific combining ability of single crosses ~N(0, D \(\sigma_{c}^{2}\)), p the vector of permanent environmental effects used to take repeated measures into account ~N(0, I \(\sigma_{p}^{2}\)), k the vector of elementary plot effects ~N(0, I \(\sigma_{k}^{2}\)) and e the vector of residual effects ~N(0, I \(\sigma_{e}^{2}\)). X, Z 1 − Z 6 are incidence matrices. A Deli and A B are matrices of additive relationships among Deli and Group B individuals, respectively, computed from pedigrees. D is the matrix of dominance relationships among crosses computed from the pedigree, with value between crosses Deli × B and Deli′ × B′ equal to f Deli,Deli′ × f B,B′ , where f Deli,Deli′ and f B,B′ are the coefficient of coancestry between the Deli and Group B parents. I is an identity matrix. For BN and ABW, the model also included a fixed age effect and a random age within cross effect a ~N(0, D \(\otimes\) I \(\sigma_{a}^{2}\)). This model was based on the model of Stuber and Cockerham (1966) for hybrids between unrelated populations, as previously used in oil palm by Purba et al. (2001). The R-ASReml package (Butler et al. 2009) for R (R Core Team 2013) was used to obtain variance component estimates and EBV of all individuals.
The accuracy of the general combining ability a i of an individual i (actually \(a_{{{\text{Deli}}_{\text{i}} }}\) or \(a_{{B_{i} }}\) depending on the population of origin of i) is given by \(r_{{a,\hat{a}_{i} }} = \sqrt {\frac{{1 - PEV_{{a_{i} }} }}{{0.5(1 + F_{i} )\sigma_{a}^{2} }}},\) where PEV ai is the prediction error variance associated with a i , 0.5(1 + F i ) is the diagonal of the relationship matrix used in the mixed model (i.e., 0.5A Deli or 0.5A B , depending on the population of origin of i), F i is the inbreeding coefficient and \(\sigma_{a}^{2}\) is the additive variance (i.e., \(\sigma_{\text{Deli}}^{2}\) or \(\sigma_{B}^{2}\), depending on the population). This formula was used to compute the mean accuracy of the general combining ability of the 131 Deli and 131 Group B parents used in the GS analysis, which was 0.89, ranging from 0.83 ± 0.06 (SD) for O/P in Deli to 0.93 ± 0.04 for K/F in Group B.
Rights and permissions
About this article
Cite this article
Cros, D., Denis, M., Sánchez, L. et al. Genomic selection prediction accuracy in a perennial crop: case study of oil palm (Elaeis guineensis Jacq.). Theor Appl Genet 128, 397–410 (2015). https://doi.org/10.1007/s00122-014-2439-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-014-2439-z