Abstract
Key message
A mixed model framework was defined for QTL analysis of multiple traits across multiple environments for a RIL population in pepper. Detection power for QTLs increased considerably and detailed study of QTL by environment interactions and pleiotropy was facilitated.
Abstract
For many agronomic crops, yield is measured simultaneously with other traits across multiple environments. The study of yield can benefit from joint analysis with other traits and relations between yield and other traits can be exploited to develop indirect selection strategies. We compare the performance of three multi-response QTL approaches based on mixed models: a multi-trait approach (MT), a multi-environment approach (ME), and a multi-trait multi-environment approach (MTME). The data come from a multi-environment experiment in pepper, for which 15 traits were measured in four environments. The approaches were compared in terms of number of QTLs detected for each trait, the explained variance, and the accuracy of prediction for the final QTL model. For the four environments together, the superior MTME approach delivered a total of 47 regions containing putative QTLs. Many of these QTLs were pleiotropic and showed quantitative QTL by environment interaction. MTME was superior to ME and MT in the number of QTLs, the explained variance and accuracy of predictions. The large number of model parameters in the MTME approach was challenging and we propose several guidelines to help obtain a stable final QTL model. The results confirmed the feasibility and strengths of novel mixed model QTL methodology to study the architecture of complex traits.








Similar content being viewed by others
Notes
The list of all abbreviations is given in Table 5 in Appendix A.
References
Alimi NA, Bink MCAM, Dieleman JA, Nicolaï M, Wubs M, Heuvelink E, Magan J, Voorrips RE, Jansen J, Rodrigues PC, Heijden GWAM, Vercauteren A, Vuylsteke M, Song Y, Glasbey C, Barocsi A, Lefebvre V, Palloix A, Eeuwijk FA (2013) Genetic and QTL analyses of yield and a set of physiological traits in pepper. Euphytica 190:181–201
Anhalt UCM, Heslop-Harrison JS, Piepho HP, Byrne S, Barth S (2009) Quantitative trait loci mapping for biomass yield traits in a Lolium inbred line derived F2 population. Euphytica 170:99–107
Barchi L, Lefebvre V, Sage-Palloix A-M, Lanteri S, Palloix A (2009) QTL analysis of plant development and fruit traits in pepper and performance of selective phenotyping. Theor Appl Genet 118:1157–1171
Barócsi A (2012) Intelligent, net or wireless enabled fluorosensors for high throughput monitoring of assorted crops. Meas Sci Technol 24:025701
Beavis WD (1994) The power and deceit of QTL experiments: Lessons from comparative QTL studies. In: Proceedings of the forty-ninth annual corn and sorghum research conference. American Seed Trade Association, Washington, pp 250–266
Beavis WD (1997) QTL analyses: power, precision, and accuracy. In: Paterson AH (ed) Molecular dissection of complex traits. CRC Press, Boca Raton, pp 145–162
Ben Chaim A, Paran I, Grube RC, Jahn M, van Wijk R, Peleman J (2001) QTL mapping of fruit-related traits in pepper (Capsicum annuum). Theor Appl Genet 102:1016–1028
Ben Chaim A, Borovsky Y, Rao G, Gur A, Zamir D, Paran I (2006) Comparative QTL mapping of fruit size and shape in tomato and pepper. Israel J Plant Sci 54:191–203
Boer MP, Wright D, Feng LZ, Podlich DW, Luo L, Cooper M, van Eeuwijk FA (2007) A mixed-model quantitative trait loci (QTL) analysis for multiple-environment trial data using environmental covariables for QTL-by-environment interactions, with an example in maize. Genetics 177:1801–1813
Hackett CA, Meyer RC, Thomas WTB (2001) Multi-trait QTL mapping in barley using multivariate regression. Genet Res 77:95–106
Jiang CJ, Zeng ZB (1995) Multiple-trait analysis of genetic-mapping for quantitative trait loci. Genetics 140:1111–1127
Kargbo A, Wang CY (2010) Complex traits mapping using introgression lines in pepper (Capsicum annuum). Afr J Agric Res 5:725–731
Klasen JR, Piepho HP, Stich B (2012) QTL detection power of multi-parental RIL populations in Arabidopsis thaliana. Heredity 108:626–632
Korte A, Vilhjalmsson BJ, Segura V, Platt A, Long Q, Nordborg M (2012) A mixed-model approach for genome-wide association studies of correlated traits in structured populations. Nat Genet 44:1066–1071
Lander ES, Botstein D (1989) Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121:185–199
Lee HR, Cho MC, Kim HJ, Park SW, Kim BD (2008) Marker development for erect versus pendant-orientated fruit in Capsicum annuum L. Mol Cells 26:548–553
Lefebvre V, Daubeze AM, van der Voort JR, Peleman J, Bardin M, Palloix A (2003) QTLs for resistance to powdery mildew in pepper under natural and artificial infections. Theor Appl Genet 107:661–666
Li J, Ji L (2005) Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity 95:221–227
MacMillan K, Emrich K, Piepho HP, Mullins CE, Price AH (2006) Assessing the importance of genotype x environment interaction for root traits in rice using a mapping population II: conventional QTL analysis. TAG 113:953–964
Malosetti M, Voltas J, Romagosa I, Ullrich SE, van Eeuwijk FA (2004) Mixed models including environmental covariables for studying QTL by environment interaction. Euphytica 137:139–145
Malosetti M, Visser RGF, Celis-Gamboa C, Eeuwijk FA (2006) QTL methodology for response curves on the basis of non-linear mixed models, with an illustration to senescence in potato. Theor Appl 113:288–300
Malosetti M, Ribaut JM, Vargas M, Crossa J, van Eeuwijk FA (2008) A multi-trait multi-environment QTL mixed model with an application to drought and nitrogen stress trials in maize (Zea mays L.). Euphytica 161:241–257
Mimura Y, Minamiyama Y, Sano H, Hirai M (2010) Mapping for axillary shooting, flowering date, primary axis length, and number of leaves in pepper (Capsicum annuum). J Jpn Soc Hortic Sci 79:56–63
Nicolaï M, Pisani C, Bouchet J, Vuylsteke M, Palloix A (2012) Discovery of a large set of SNP and SSR genetic markers by high-throughput sequencing of pepper (Capsicum annuum). Genet Mol Res 11:2295–2300
Panozzo JF, Eckermann PJ, Mather DE, Moody DB, Black CK, Collins HM, Barr AR, Lim P, Cullis BR (2007) QTL analysis of malting quality traits in two barley populations. Aust J Agric Res 58:858–866
Piepho H-P (2000) A mixed-model approach to mapping quantitative trait loci in barley on the basis of multiple environment data. Genetics 156:2043–2050
Rao GU, Ben Chaim A, Borovsky Y, Paran I (2003) Mapping of yield-related QTLs in pepper in an interspecific cross of Capsicum annuum and C. frutescens. Theor Appl Genet 106:1457–1466
Sukhwinder S, Hernandez MV, Crossa J, Singh PK, Bains NS, Singh K, Sharma I (2012) Multi-trait and multi-environment QTL analyses for resistance to wheat diseases. PLoS One 7:e38008
van der Heijden G, Song Y, Horgan G, Polder G, Dieleman A, Bink M, Palloix A, van Eeuwijk F, Glasbey C (2012) SPICY: towards automated phenotyping of large pepper plants in the greenhouse. Funct Plant Biol 39:870–877
van Eeuwijk FA (ed) (2012) Smart tools for prediction and improvements of crop yield—KBBE 211347. http://www.spicyweb.eu
van Eeuwijk FA, Bink M, Chenu K, Chapman SC (2010) Detection and use of QTL for complex traits in multiple environments. Curr Opin Plant Biol 13:193–205
Vargas M, van Eeuwijk F, Crossa J, Ribaut J-M (2006) Mapping QTLs and QTL × environment interaction for CIMMYT maize drought stress program using factorial regression and partial least squares methods. Theor Appl Genet 112:1009–1023
Verbeke G, Molenberghs G (2000) Linear mixed models for longitudinal data. Springer, New York
Verbyla AP, Eckermann PJ, Thompson R, Cullis BR (2003) The analysis of quantitative trait loci in multi-environment trials using a multiplicative mixed model. Aust J Agric Res 54:1395–1408
Vilhjalmsson BJ, Nordborg M (2013) The nature of confounding in genome-wide association studies. Nat Rev Genet 14:1–2
Voorrips RE, Palloix A, Dieleman JA, Bink MCAM, Heuvelink E, Heijden GWAM, van der Vuylsteke M, Glasbey C, Barócsi A, Magán J, van Eeuwijk FA (2010) Crop growth models for the -omics era: the EU-SPICY project. In: Advances in genetics and breeding of capsicum and eggplant: Proceedings of the XIVth EUCARPIA meeting on genetics and breeding of capsicum and eggplant. Editorial Universidad Politécnica de Valencia, Valencia, pp 315–321
VSNi (2012) GenStat for Windows 15th Edition. VSN International, Hemel Hempstead
Weller J, Soller M (2004) An analytical formula to estimate confidence interval of QTL location with a saturated genetic map as a function of experimental design. Theor Appl Genet 109:1224–1229
West-Eberhard MJ (2003) Developmental plasticity and evolution. Oxford University Press, New York
Xu S (2013) Mapping QTL for multiple traits. In: Principles of statistical genomics. Springer, New York, pp 209–222
Zeng ZB (1994) Precision mapping of quantitative trait loci. Genetics 136:1457–1468
Zygier S, Chaim AB, Efrati A, Kaluzky G, Borovsky Y, Paran I (2005) QTLs mapping for fruit size and shape in chromosomes 2 and 4 in pepper and a comparison of the pepper QTL map with that of tomato. Theor Appl Genet 111:437–445
Acknowledgments
The research leading to these results has received funding from the European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement n° 211347. We thank the EU-SPICY Industrial Advisory Board for support and discussions. Rik van Wijk and Syngenta are especially acknowledged for their highly valuable help in making available additional SNP markers that strongly improved the quality of the genetic map. Roeland Voorrips and other members of the EU-SPICY project are acknowledged for their contributions and helpful comments. We also thank Paul Keizer, Marcos Malosetti and Martin Boer of Biometris for their valuable insights.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standard
The authors declare that the experiments in this study comply with the current laws of the countries (Spain and Netherlands) in which the experiments were performed.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by I. Mackay.
Appendices
Appendix A
Appendix B
See Fig. 9.
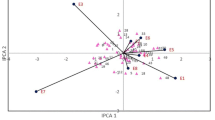
Biplots for BLUEs and fitted trait values in each environment. NL1, NL2, SP1 and SP2 are the biplots of BLUE for the traits in each environment while NL1p, NL2p, SP1p and SP2p are the biplots for fitted values of each trait in each environment from the MTME QTL model. The cosine of the angle between the lines approximates the correlation between the traits they represent. The closer the angles are, the higher the correlations. Angles close to 90 or 270° reflect weaker correlations. In each environment, angles between traits are similar for biplots from BLUEs and fitted values. E.g. the biplot for NL1 and NL1p, show a strong relationship between DWF and NF, and a weak relationship between DWF and NLE. The lines enclosing the sample points in the biplots are known as convex hulls, representing the smallest convex set of the sample data
Appendix C: QTL by environment results from ME analyses
CIM Profile plot for all the traits in the multi-environment analyses. The top section shows the P-values of tests for QTL main effects. The bottom section shows heat maps along the genome for each environment, where blue means that the YW allele had a significant positive effect and red means that the CM334 allele had a significant positive effect in that environment. Many of the QTLs are constitutive i.e. consistent across environments with no crossover interaction except the QTL on chromosome 11 for LUE, Axl, SL and INL (colour figure online)
Appendix D: QTL effects from MT analyses
See Table 9.
Appendix E: QTL effects from MTME analyses: chromosomes 3–12
Rights and permissions
About this article
Cite this article
Alimi, N.A., Bink, M.C.A.M., Dieleman, J.A. et al. Multi-trait and multi-environment QTL analyses of yield and a set of physiological traits in pepper. Theor Appl Genet 126, 2597–2625 (2013). https://doi.org/10.1007/s00122-013-2160-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-013-2160-3