Abstract
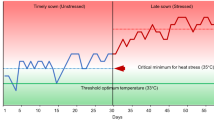
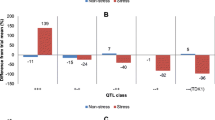
Short-term submergence is a recurring problem in many rice production areas. The SUB1 gene, derived from the tolerant variety FR13A, has been transferred to a number of widely grown varieties, allowing them to withstand complete submergence for up to 2 weeks. However, in areas where longer-term submergence occurs, improved varieties having higher tolerance levels are needed. To search for novel quantitative trait loci (QTLs) from other donors, an F2:3 population between IR72 and Madabaru, both moderately tolerant varieties, was investigated. After a repeated phenotyping of 466 families under submergence stress, a subset of 80 families selected from the two extreme phenotypic tails was used for the QTL analysis. Phenotypic data showed transgressive segregation, with several families having an even higher survival rate than the FR13A-derived tolerant check (IR40931). Four QTLs were identified on chromosomes 1, 2, 9, and 12; the largest QTL on chromosome 1 had a LOD score of 11.2 and R 2 of 52.3%. A QTL mapping to the SUB1 region on chromosome 9, with a LOD score of 3.6 and R 2 of 18.6%, had the tolerant allele from Madabaru, while the other three QTLs had tolerant alleles from IR72. The identification of three non-SUB1 QTLs from IR72 suggests that an alternative pathway may be present in this variety that is independent of the ethylene-dependent pathway mediated by the SUB1A gene. These novel QTLs can be combined with SUB1 using marker assisted backcrossing in an effort to enhance the level of submergence tolerance for flood-prone areas.




Similar content being viewed by others
References
Clark RM, Wagler TN, Quijada P, Doebley J (2006) A distant upstream enhancer at the maize domestication gene tb1 has pleiotrophic effects on plant and inflorescent architecture. Nat Genet 38:594–597
Fukao T, Bailey-Serres J (2008) Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellins responses in rice. Proc Natl Acad Sci USA 105:16814–16819
Iftekharuddaula KM, Newaz MA, Salam MA, Ahmed HU, Mahbub MAA, Septiningsih EM, Collard BCY, Sanchez DL, Pamplona AM, Mackill DJ (2011) Rapid and high-precision marker assisted backcrossing to introgress the SUB1 QTL into BR11, the rainfed lowland rice mega variety of Bangladesh. Euphytica 178:83–97
Khush GS, Virk PS (2005) IR varieties and their impact. International Rice Research Institute, Los Baños, p 163 (Philippines)
Li Z-X, Septiningsih EM, Quiloy-Mercado SM, McNally KL, Mackill DJ (2010) Identification of SUB1A alleles from wild rice Oryza rufipogon Griff. Genet Resour Crop Evol 58:1237–1242
Manly KF, Cudmore RH Jr, Meer JM (2001) Map Manager QTX, cross-platform software for genetic mapping. Mamm Genome 12:930–932
McCouch SR, Sweeney M, Li J, Jiang H, Thomson M, Septiningsih E, Edwards J, Moncada P, Xiao J, Garris A, Tai T, Martinez C, Tohme J, Sugiono M, McClung A, Yuan LP, Ahn SN (2007) Through the genetic bottleneck: O. rufipogon as a source of trait-enhancing alleles for O. sativa. Euphytica 154:317–339
McCouch SR (2008) Gene nomenclature system for rice. Rice 1:72–84
Nandi S, Subudhi PK, Senadhira D, Manigbas NL, Sen-Mandi S, Huang N (1997) Mapping QTLs for submergence tolerance in rice by AFLP analysis and selective genotyping. Mol Gen Genet 255:1–8
Neeraja C, Maghirang-Rodriguez R, Pamplona A, Heuer S, Collard B, Septiningsih E, Vergara G, Sanchez D, Xu K, Ismail A, Mackill D (2007) A marker-assisted backcross approach for developing submergence-tolerant rice cultivars. Theor Appl Genet 115:767–776
Nelson JC (1997) QGENE: software for marker-based genomic analysis and breeding. Mol Breed 3:239–245
Pallotta MA, Graham RD, Langridge P, Sparrow DHB, Barker SJ (2000) RFLP mapping of manganese efficiency in barley. Theor Appl Genet 101:1100–1108
Sarkar RK, Panda D, Reddy JN, Patnaik SSC, Mackill DJ, Ismail AM (2009) Performance of submergence tolerant rice (Oryza sativa) genotypes carring the Sub1 quantitative trait locus under stressed and non-stressed natural field conditions. Indian J Agric Sci 79:876–883
Septiningsih EM, Pamplona AM, Sanchez DL, Neeraja CN, Vergara GV, Heuer S, Ismail AM, Mackill DJ (2009) Development of submergence tolerant rice cultivars: the Sub1 locus and beyond. Ann Bot 103:151–160
Septiningsih EM, Prasetiyono J, Lubis E, Tai TH, Tjubaryat T, Moeljopawiro S, McCouch SR (2003) Identification of quantitative trait loci for yield and yield components in an advanced backcross population derived from the Oryza sativa variety IR64 and the wild relative O. rufipogon. Theor Appl Genet 107:1419–1432
Singh N, Dang TTM, Vergara GV, Pandey DM, Sanchez D, Neeraja CN, Septiningsih EM, Mendioro M, Tecson-Mendoza EM, Ismail AM, Mackill DJ, Heuer S (2010) Molecular marker survey and expression analyses of the rice submergence-tolerance genes SUB1A and SUB1C. Theor Appl Genet 121:1441–1453
Singh S, Mackill DJ, Ismail AM (2009) Responses of SUB1 rice introgression lines to submergence in the field: yield and grain quality. Field Crop Res 113:12–23
Thomson MJ, Tai TH, McClung AM, Hinga ME, Lobos KB, Xu Y, Martinez C, McCouch SR (2003) Mapping quantitative trait loci for yield, yield components, and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor Appl Genet 107:479–493
Toojinda T, Siangliw M, Tragroonrung S, Vanavichit A (2003) Molecular genetics of submergence tolerance in rice: QTL analysis of key traits. Ann Bot 91:243–253
Wang S, Basten CJ, Zeng Z-B (2010) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC. http://statgen.ncsu.edu/qtlcart/WQTLCart.htm
Xu K, Mackill DJ (1996) A major locus for submergence tolerance mapped on rice chromosome 9. Mol Breed 2:219–224
Xu K, Xia X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ (2006) Sub1A is an ethylene response factor-like gene that confers submergence tolerance to rice. Nature 442:705–708
Xu K, Xu X, Ronald PC, Mackill DJ (2000) A high-resolution linkage map in the vicinity of the rice submergence tolerance locus SUB1. Mol Gen Genet 263:681–689
Yang J, Zhu J, Williams RW (2007) Mapping the genetic architecture of complex traits in experimental populations. Bioinformatics 23:1527–1536
Yang J, Hu CC, Hu H, Yu RD, Xia Z, Ye XZ, Zhu J (2008) QTLNetwork: mapping and visualizing genetic architecture of complex traits in experimental populations. Bioinformatics 24:721–723
Yang J, Zhu J (2005) Methods for predicting superior genotypes under multiple environments based on QTL effects. Theor Appl Genet 110:1268–1274
Zheng K, Subudhi PK, Domingo J, Magpantay G, Huang N (1995) Rapid DNA isolation for marker assisted selection in rice breeding. Rice Genet Newslett 12:255–258
Acknowledgments
Technical assistance from R. Garcia, E. Suiton, J. Mendoza, J. C. Ignacio, A. S. M. Masuduzzaman, and V. Bartolome are gratefully acknowledged. The work reported here was supported in part by a grant from the Bill and Melinda Gates Foundation (BMGF) through the project on “Stress-tolerant rice for Africa and South Asia” (STRASA).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by L. Xiong.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Septiningsih, E.M., Sanchez, D.L., Singh, N. et al. Identifying novel QTLs for submergence tolerance in rice cultivars IR72 and Madabaru. Theor Appl Genet 124, 867–874 (2012). https://doi.org/10.1007/s00122-011-1751-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-011-1751-0