Abstract
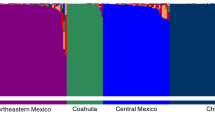
Using 20 SSR markers well scattered across the 19 grape chromosomes, we analyzed 4,370 accessions of the INRA grape repository at Vassal, mostly cultivars of Vitis vinifera subsp. sativa (3,727), but also accessions of V. vinifera subsp. sylvestris (80), interspecific hybrids (364), and rootstocks (199). The analysis revealed 2,836 SSR single profiles: 2,323 sativa cultivars, 72 wild individuals (sylvestris), 306 interspecific hybrids, and 135 rootstocks, corresponding to 2,739 different cultivars in all. A total of 524 alleles were detected, with a mean of 26.20 alleles per locus. For the 2,323 cultivars of V. vinifera, 338 alleles were detected with a mean of 16.9 alleles per locus. The mean genetic diversity (GDI) was 0.797 and the level of heterozygosity was 0.76, with broad variation from 0.20 to 1. Interspecific hybrids and rootstocks were more heterozygous and more diverse (GDI = 0.839 and 0.865, respectively) than V. vinifera cultivars (GDI = 0.769), Vitis vinifera subsp. sylvestris being the least divergent with GDI = 0.708. Principal coordinates analysis distinguished the four groups. Slight clonal polymorphism was detected. The limit between clonal variation and cultivar polymorphism was set at four allelic differences out of 40. SSR markers were useful as a complementary tool to traditional ampelography for cultivar identification. Finally, a set of nine SSR markers was defined that was sufficient to distinguish 99.8% of the analyzed accessions. This set is suitable for routine characterization and will be valuable for germplasm management.




Similar content being viewed by others
References
Adam-Blondon AF, Roux C, Claux D, Butterlin G, Merdinoglu D, This P (2004) Mapping 245 SSR markers on the Vitis vinifera genome: a tool for grape genetics. Theor Appl Genet 109:1017–1027
Allcochete AAN, Rangel PHN, Ferreira ME (2008) Genetic structure of rices samples from germplasm bank. Sci Res Essays 3:577–598
Aradhya MK, Dangl GS, Prins BH et al (2003) Genetic structure and differentiation in cultivated grape, Vitis vinifera L. Genet Res 81:179–192
Arnold C, Rossetto M, McNally J, Henry RJ (2002) The application of SSRs characterized for grape (Vitis vinifera) to conservation studies in Vitaceae. Am J Bot 89:22–28
Bacilieri R (2007) GrapeGen06-management and conservation of grapevine GR. Bioversity Newsl Eur 34:16
Boursiquot J-M, This P (1996) Les nouvelles techniques utilisées en ampélographie: informatique et marquage. J Int Sci Vigne Vin (special issue) 13–23
Boursiquot J-M, This P (2000) Essai de définition du cépage. Revue des Œnologues 94:5–7
Boursiquot J-M, Lacombe T, Laucou V, Julliard S, Perrin F-X, Lanier N, Legrand D, Meredith C, This P (2009) Parentage of Merlot and related winegrape cultivars of southwestern France: discovery of the missing link. Aust J Grape Wine Res 15:144–155
Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL (1994) High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368:455–457
Bowers JE, Dangl GS, Vignani R, Meredith CP (1996) Isolation and characterization of new polymorphic simple sequence repeat loci in grape (Vitis vinifera L.). Genome 39:628–633
Bowers JE, Dangl GS, Meredith CP (1999a) Development and characterization of additional microsatellite DNA markers for grape. Am J Enol Vitic 50:243–246
Bowers JE, Boursiquot J-M, This P, Chu K, Johansson H, Meredith C (1999b) Historical genetics: the parentage of chardonnay, gamay, and other wine grapes of northeastern France. Science 285:1562–1565
Cipriani G, Marrazzo MT, Di Gaspero G, Pfeiffer A, Morgante M, Testolin R (2008) A set of microsatellite markers with long core repeat optimized for grape (Vitis spp.) genotyping. BMC Plant Biol 8:127
Cipriani G, Spadotto A, Jurman I, Di Gaspero G, Crespan M, Meneghetti S, Frare E, Vignani R, Cresti M, Morgante M, Pezzotti M, Pe E, Policriti A, Testolin R (2010) The SSR-based molecular profile of 1005 grapevine (Vitis vinifera L.) accessions uncovers new synonymy and parentages, and reveals a large admixture amongst varieties of different geographic origin. Theor Appl Genet 121:1569–1585
Dangl GS, Mendum ML, Prins BH, Walker MA, Meredith CP, Simon CJ (2001) Simple sequence repeat analysis of a clonally propagated species: a tool for managing a grape germplasm collection. Genome 44:432–438
De Andrès MT, Cabezas JA, Cerveza MT, Borrego J, Martinèz-Zapater JM, Jouve N (2007) Molecular characterization of grapevine rootstocks maintained in germplasm collections. Am J Enol Vitic 58:75–86
Di Gaspero G, Peterluner E, Testolin R, Edwards KJ, Cipriani G (2000) Conservation of microsatellite loci within the genus Vitis. Theor Appl Genet 101:301–308
Di Vecchi-Staraz M, This P, Boursiquot J-M, Laucou V, Lacombe T, Bandinelli R, Varès D, Boselli M (2007) Genetic structuring and parentage analysis for evolutionary studies in grapevine: kingroup and origin of cv. Sangiovese revealed. J Am Soc Hortic Sci 132:514–524
Di Vecchi-Staraz M, Laucou V, Bruno G, Lacombe T, Gerber S, Bourse T, Boselli M, This P (2009) Low level of pollen-mediated gene flow from cultivated to wild grapevine: consequences for the evolution of the endangered subspecies Vitis vinifera L. subsp. silvestris. J Hered 100:66–75
Doligez A, Adam-Blondon AF, Cipriani G, Di Gaspero G, Laucou V, Merdinoglu D, Meredith CP, Riaz S, Roux C, This P (2006) An integrated SSR map of grapevine based on five mapping populations. Theor Appl Genet 113:369–382
Duchêne E, Butterlin G, Claudel P, Dumas V, Jaegli N, Merdinoglu D (2009) A grapevine (Vitis vinifera L.) deoxy-d-xylulose synthase gene colocates with a major quantitative trait loci for terpenol content. Theor Appl Genet 118:541–552
Esselink GD, Smulders MJM, Vosman B (2003) Identification of cut rose (Rosa hybrida) and rootstock varieties using robust sequence tagged microsatellite site markers. Theor Appl Genet 106:277–286
Fernandez-Gonzalez M, Mena A, Izquierdo P, Martinez J (2007) Genetic characterization of grapevine (V. vinifera L.) cultivars from Castilla La Mancha (Spain) using microsatellite markers. Vitis 46:126–130
Franks T, Botta R, Thomas MR (2002) Chimerism in grapevines: implications for cultivar identity, ancestry, and genetic improvement. Theor Appl Genet 104:192–199
Galet P (2000) Dictionnaire encyclopédique des cépages. Hachette Livre
Gao LZ, Zhang CH, Li DY, Pan DJ, Jia JZ, Dong YS (2006) Genetic diversity within Oryza rufipogon germplasms preserved in Chinese field gene banks of wild rice as revealed by microsatellite markers. Biodivers Conserv 15:4059–4077
Gerber S, Chabrier P, Kremer A (2003) FAMOZ: a software for parentage analysis using dominant, codominant and uniparentaly inherited markers. Mol Ecol 3:479–481
Giannetto S, Velasco R, Troggio M, Malacarne G, Storchi P, Cancellier S, De Nardi B, Crespan M (2008) A PCR-based diagnostic tool for distinguishing grape skin color mutants. Plant Sci 175:402–409
Grassi F, Labra M, Imazio S, Spada A, Sgorbati S, Scienza A, Sala F (2003) Evidence of a secondary grapevine domestication center detected by SSR analysis. Theor Appl Genet 107:1315–1320
Gupta PK, Varshney RK (2000) The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphytica 113:163–185
Ibanez J, Velez MD, de Andres MT, Borrego J (2009) Molecular markers for establishing distinctness in vegetatively propagated crops: a case study in grapevine. Theor Appl Genet 119:1213–1222
Imazio S, Labra M, Grassi F, Winfield M, Bardini M, Scienza A (2002) Molecular tools for clone identification: the case of the grapevine cultivar ‘traminer”. Plant Breed 121:531–535
Jaillon O, Aury J-M, Noel B, Policriti A, Clepet C, Casagrande A et al (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449(7161):463–467
Kloosterman AD, Budowle B, Daselaar P (1993) PCR-amplification and detection of the human D1S80 VNTR locus. Int J Legal Med 105:257–264
Koo B, Pardey PG, Wright BD (2004) Saving seeds: the economics of conserving crop genetic resources ex situ in the future harvest centres of CGIAR. Bioversity International, Rome, Italy
Le Cunff L, Fournier-Level A, Laucou V, Vezzulli S, Lacombe T, Adam-Blondon A-F, Boursiquot J-M, This P (2008) Construction of nested genetic core collections to optimize the exploitation of natural diversity in V. vinifera L. subsp sativa. BMC Plant Biol 8:31
Levadoux L, Boubals D, Rives M (1962) Le genre Vitis et ses espèces. Ann Amelior Plant 12:19–44
Martinez LE, Cavagnaro PF, Masuelli RW, Zuniga M (2006) SSR-based assessment of genetic diversity in South American Vitis vinifera varieties. Plant Sci 170:1036–1044
Maul E, This P (2008) GENRES 081—a basis for the preservation and utilization of Vitis genetic resources. In: Report of a working group on Vitis. First meeting, 12–14 June 2003, Palić, Serbia and Montenegro. Bioversity International, Rome, Italy, pp 13–22
McGovern PE (2003) Ancient wine. The search for the origins of viniculture. Princeton University Press, Princeton, NJ
Merdinoglu D, Butterlin G, Bevilacqua L, Chiquet V, Adam-Blondon A-F, Decroocq S (2005) Development and characterization of a large set of microsatellite markers in grapevine (Vitis vinifera L.) suitable for multiplex PCR. Mol Breed 15:349–366
Myles S, Chia JM, Hurwitz B, Simon C, Zhong GY, Buckler E, Ware D (2010) Rapid genomic characterization of the genus Vitis. PLoS ONE 5(1):e8219. doi:10.1371/journal.pone.0008219
Park SDE (2001) Trypanotolerance in West African cattle and the population genetic effects of selection. PhD thesis, University of Dublin
Pelsy F (2007) Untranslated leader region polymorphism of Tvv1, a retrotransposon family, is a novel marker useful for analyzing genetic diversity and relatedness in the genus Vitis. Theor Appl Genet 116:15–27
Peng JH, Bai Y, Haley SD, Lapitan NLV (2009) Microsatellite-based molecular diversity of bread wheat germplasm and association mapping of wheat resistance to the Russian wheat aphid. Genetica 135:95–122
Perrier X, Jacquemoud-Collet J-P (2006) DARwin software. http://darwin.cirad.fr/darwin
Rafalski A (2002) Applications of single nucleotide polymorphisms in crop genetics. Curr Opin Plant Biol 5:94–100
Ranc N, Munos S, Santoni S, Causse M (2008) A clarified position for Solanum lycopersicum var. cerasiforme in the evolutionary history of tomatoes (Solanaceae). BMC Plant Biol 8:130
Santana JC, Heuertz M, Arranz C, Rubio JA, Martinez-Zapater JM, Hidalgo H (2010) Genetic structure, origins, and relationships of grapevine cultivars from the Castilian plateau of Spain. Am J Enol Vitic 61:214–224
Schellenbaum P, Mohler V, Wenzel G, Walter B (2008) Variation in DNA methylation patterns of grapevine somaclones (Vitis vinifera L.). BMC Plant Biol 8:78
Sefc KM, Lopes MS, Lefort F, Botta R, Roubelakis-Angelakis KA, Ibañez J, Pejic I, Wagner HW, Glössl J, Steinkellner H (2000) Microsatellite variability in grapevine cultivars from different European regions and evaluation of assignment testing to assess the geographic origin of cultivars. Theor Appl Genet 100:498–505
Sefc KM, Lefort F, Grando MS, Scott K, Steinkellner H, Thomas MR (2001) Microsatellite markers for grapevine: a state of the art. In: Roubelakis-Angelakis KA (ed) Molecular biology biotechnology of grapevine. Kluwer Publishers, Amsterdam, pp 433–463
Smulders MJM, Cottrell JE, Le fever F, van der Shoot J, Arens P, Vosman B et al (2008) Structure of the genetic diversity in black poplar (Populus nigra L.) populations across European river systems: consequences for conservation and restoration. For Ecol Manag 255:1388–1399
This P, Roux C, Parra P, Siret R, Bourse T, Adam A-F, Yvon M, Lacombe T, David J, Boursiquot J-M (2000) Caractérisation de la diversité d’une population de vignes sauvages du Pic Saint Loup (Hérault) et relations avec le compartiment cultivé. Genet Sel Evol 33:S289–S304
This P, Jung A, Boccacci P, Borrego J, Botta R, Costantini L et al (2004) Development of a standard set of microsatellite reference alleles for identification of grape varieties. Theor Appl Genet 109:1448–1458
This P, Lacombe T, Thomas MR (2006) Historical origins and genetic diversity of wine grapes. Trends Genet 22:511–519
This P, Cadle-Davidson M, Lacombe T, Owens CL (2007) Wine grape (Vitis vinifera L.) color associates with allelic variation in the domestication gene VvmybA1. Theor Appl Genet 114:723–730
Thomas MR, Scott NS (1993) Microsatellite repeats in grapevine reveal DNA polymorphisms when analyzed as sequence-tagged sites (STSS). Theor Appl Genet 86:985–990
Thomas MR, Matsumotio S, Cain P, Scott NS (1993) Repetitive DNA of grapevine: classes present and sequences suitable for cultivar identification. Theor Appl Genet 86:173–180
Vezzulli S, Micheletti D, Riaz S, Pindo M, Viola R, This P, Walker A, Troggio M, Velasco R (2008) A SNP transferability survey within the genus Vitis. BMC Plant Biol 8:128
Vigouroux Y, Glaubitz JC, Matsuoka Y, Goodman MM, Sanchez JG, Doebley J (2008) Population structure and genetic diversity of new world maize races assessed by DNA microsatellites. Am J Bot 95:1240–1253
Weir BS (1989) Sampling properties of gene diversity. In: Brown AHD, Clegg MT, Kahler HL, Weir BS (eds) Plant population genetics, breeding, genetic ressources. Sinauer Associates, Sunderland, MA, pp 23–42
Zoghlami N, Riahi L, Laucou V, Lacombe T, Mliki A, Ghorbel A, This P (2009) Origin and genetic diversity of Tunisian grapes as revealed by microsatellite markers. Sci Hortic 120:479–486
Acknowledgment
This work was supported by a grant from Ministère de l’Agriculture, de la Pêche et de la forêt (Centre de Ressources Biologiques).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Frisch.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Laucou, V., Lacombe, T., Dechesne, F. et al. High throughput analysis of grape genetic diversity as a tool for germplasm collection management. Theor Appl Genet 122, 1233–1245 (2011). https://doi.org/10.1007/s00122-010-1527-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-010-1527-y