Abstract
The Grp1 locus confers broad-spectrum resistance to the potato cyst nematode species Globodera pallida and Globodera rostochiensis and is located in the GP21-GP179 interval on the short arm of chromosome V of potato. A high-resolution map has been developed using the diploid mapping population RHAM026, comprising 1,536 genotypes. The flanking markers GP21 and GP179 have been used to screen the 1,536 genotypes for recombination events. Interval mapping of the resistances to G. pallida Pa2 and G. rostochiensis Ro5 resulted in two nearly identical LOD graphs with the highest LOD score just north of marker TG432. Detailed analysis of the 44 recombinant genotypes showed that G. pallida and G. rostochiensis resistance could not be separated and map to the same location between marker SPUD838 and TG432. It is suggested that the quantitative resistance to both nematode species at the Grp1 locus is mediated by one or more tightly linked R genes that might belong to the NBS-LRR class.
Similar content being viewed by others
Introduction
The potato cyst nematode (PCN) species Globodera pallida and Globodera rostochiensis cause serious yield losses in potato crops worldwide (Oerke et al. 1994). PCN can be controlled by crop rotation, chemical soil disinfestations and the use of resistant cultivars. However, due to the formation of cysts, PCN can survive in the soil for many years in the absence of a host, making crop rotation unattractive for potato farmers. Chemical control of PCN involves very unspecific and extremely harmful pesticides. Due to increasing concern about environmental issues and governmental regulations, this method has been practically abandoned in many countries. Therefore, resistant cultivars are becoming increasingly important and, hence, scientific studies on the underlying genes and resistance mechanisms are of great interest.
A total of 14 PCN resistance loci have been mapped in potato on chromosomes III, IV, V, VII, IX, X, XI and XII (reviewed by Gebhardt and Valkonen 2001; Caromel et al. 2003, 2005. Ten resistance traits confer partial resistance (Gro1.4, Gpa4, Gpa, Gpa5, Grp1, Gpa6, Gro1.2, Gro1.3 and GpaM1), while four of them (H1, GroVI, GroI and Gpa2) and the combination of GpaV s spl and GpaXI s spl confer nearly absolute resistance to one or more pathotypes. Many of these PCN resistance loci are mapped in regions of the potato genome where clusters of resistance gene homologues are located. This is not only true for the single dominantly inherited PCN resistance (R) genes Gpa2 and Gro1 (Barone et al. 1990; Rouppe van der Voort et al. 1997), but also for quantitative trait loci (QTL) such as Grp1, Gpa, GpaV spl , GpaM1 and Gpa5 (Caromel et al. 2003, 2005; Kreike et al. 1994; Rouppe van der Voort et al. 1998, 2000). So far, the only nematode R genes that have been characterised at the molecular level in potato are Gpa2 and Gro1 (Paal et al. 2004; Van der Vossen et al. 2000).
Resistance conferred by the Grp1 locus was discovered in 1998 in the tetraploid clone AM78-3778, an interspecific hybrid between S. tuberosum and several wild potato species including S. vernei, S. oplocense and S. tuberosum ssp. andigena (Rouppe van der Voort et al. 1998). This locus is particularly interesting since it confers major resistance to the potato cyst nematode (PCN) pathotypes Ro5 (G. rostochiensis line 22) and Pa2 (G. pallida population D383), as well as partial resistance to pathotype Pa3 (G. pallida population Rookmaker). The Grp1 locus has been mapped in the GP21-GP179 interval on the short arm of chromosome V of the diploid potato clone 3778-16, which is derived from AM78-3778 (Rouppe van der Voort et al. 1998). This region is known to harbour resistance loci with specificities to different plant pathogens, including R1 (Leonards-Schippers et al. 1992) and a major QTL (Leonards-Schippers et al. 1994) to Phytophthora infestans, Rx2 (Ritter et al. 1991) and Nb (De Jong et al. 1997) to potato virus X, as well as Gpa5 (Rouppe van der Voort et al. 2000), Gpa (Kreike et al. 1994), GpaM1 (Caromel et al. 2003) and GpaV s spl (Caromel et al. 2005) to G. pallida. A QTL involved in trichome-mediated insect resistance has also been detected in this region (Bonierbale et al. 1994). R1 and Rx2 have been shown to belong to the nucleotide binding site leucine-rich repeat (NBS-LRR) superfamily of R genes (Ballvora et al. 2002; Bendahmane et al. 2000).
The broad-spectrum resistance of Grp1 together with its location in a hotspot of resistance led to the hypothesis that Grp1 is a compound locus containing different R genes for PCN resistance (Rouppe van der Voort et al. 1998). This is strengthened by the fact that AM78-3778 is the result of many generations of breeding, involving different wild Solanum sources (Dellaert and Vinke 1987) and by the detection of the G. pallida resistance locus Gpa5 in clone 3704-76, which is very closely related to 3778-16 (Rouppe van der Voort et al. 2000). The use of common markers for the mapping of the QTL for both Grp1 and Gpa5 showed that the two QTLs perfectly superimpose on each other, which indicates the presence of similar introgression segments. Since G. pallida resistance has been an important trait for the selection of both breeding lines, it is argued that the same gene(s) underlie(s) the G. pallida resistance. It is assumed that the G. rostochiensis resistance may have been lost during the course of the breeding process due to a recombination between loci TG432 and GP179 in a progenitor of clone 3704-76.
In this study, a high-resolution map of the Grp1 locus has been constructed, using QTL analysis. The resulting high resolution map shows similar QTLs for both G. rostochiensis and G. pallida resistance close to marker TG432. After classification of the genotypes for resistance to G. pallida Pa2 and G. rostochiensis Ro5, Grp1 resistance behaved as a monogenic R gene and could be mapped between the markers SPUD839 and TG432. The segregating alleles of the two flanking markers are in coupling phase with and tightly linked to the Grp1 locus (0.9 and 0.2 cM, respectively). Together with information on candidate genes in the area, this will form the basis for the identification of the gene(s) underlying Grp1 resistance to populations of both G. pallida and G. rostochiensis.
Materials and methods
Plant material
A population (F1AMRH) of 1,536 F1 genotypes from the cross between the diploid potato clones 3778-16 (AM) × RH89-039-16 (RH) were used (Park et al. 2005). The female parent (AM) harbours the Grp1 locus that confers major resistance to the PCN pathotypes Ro5 (G. rostochiensis line 22) and Pa2 (G. pallida population D383), as well as partial resistance to pathotype Pa3 (G. pallida population Rookmaker) and was produced by prickle pollination of the tetraploid potato clone AM 78-3778 with haploid S. phureja inducer clones (Rouppe van der Voort et al. 1998). AM78-3778 is an interspecific hybrid between S. tuberosum and several wild potato species including S. vernei 24/20, S. vernei ssp. ballsii 2/1, S. vernei LGU 8, S. oplocense EBS 1786 and S. tuberosum ssp. andigena CPC 1673. The male parent (RH) is fully susceptible to all potato cyst nematode populations tested.
Marker analysis
Genomic DNA from AM, RH and progeny was available (Park et al. 2005). CAPS markers GP21 and GP179 were used as described (Rouppe van der Voort et al. 1998). Thirteen PCR markers were designed based on sequence information derived from chromosome V of S. demissum or from the GABI database (Riano-Pachon et al. 2009). The DNA sequences of the PCR primers, their background and the corresponding thermal cycling conditions used for each are presented in Table 1. Five primer combinations resulted in AM allele specific PCR markers. The remaining eight primer combinations resulted in segregating AM alleles after digestion of the amplification products using indicated restriction endonuclease (Table 1). Primer sequences of markers SPUD839, TG432 and SPUD1636 were as described (Bryan et al. 2002; De Jong et al. 1997).
Resistance assays
PCN resistance assays were performed on plants derived from tubers as described (Rouppe van der Voort et al. 1997). The inheritance of resistance to PCN populations Ro5-22 and Pa2-D383 was analysed in population F1AMRH using three replicates of genotypes that showed a recombination event between markers GP21 and GP179. Cv Eigenheimer was included as a susceptible standard.
Data analysis and QTL mapping
Analysis of variance was carried out on log10(x + 1) transformed average cyst counts per plant genotype according to the model:
where σ 2tot is the phenotypic variance, σ 2gen is the genetic variance among the plant genotypes and σ 2rep is the environmental variance among replications.
Broad-sense heritability was estimated according to the formula:
where σ 2gen is the genetic variance among the plant genotypes, σ 2 e is the error variance and n is the number of replicates.
The data on marker segregation of the resistant parent AM was included for QTL analysis using the program MapQTL5 (Van Ooijen 2004). An LOD value of 2.5 was chosen as a threshold value (Lander and Botstein 1989).
Results
Markers closely linked to the Grp1 locus
The nematode resistance locus Grp1 has previously been mapped in a 3-cM interval on chromosome V of the diploid potato clone AM flanked by CAPS markers GP21 and GP179 (Rouppe van der Voort et al. 1998). To increase the resolution in this interval, 1,536 progeny of the mapping population RHAM026 were screened for the presence or absence of the markers GP21 and GP179 (Table 1). A total of 61 genotypes showed a recombination event between these two markers, resulting in an interval of 3.97 cM.
To identify markers closely linked to the Grp1 locus, primers were designed on sequences derived from this locus in other potato genotypes that were retrieved from Genbank and the GABI database. Thirteen primer combinations resulted in a segregating the AM allele. Table 1 presents the markers in more detail. Five markers were AM allele specific, showing the presence or absence of the amplification product. Eight markers were polymorphic after digestion with an appropriate enzyme (CAPS markers). In addition, polymorphisms were found for markers SPUD839 and TG432 (De Jong et al. 1997) and for SPUD1636 (Bryan et al. 2002). The 16 markers that revealed a polymorphism between AM and RH were subsequently tested on a subset of 44 genotypes that showed a recombination event in the GP21–GP179 interval. All markers were placed between GP21 and GP179 (Fig. 1). Some markers were not separated by recombination events and were grouped together.
Fine mapping of the GP21–GP179 interval on chromosome V of the diploid potato clone AM. Markers are presented in the correct genetic order. SPUD839, TG432, BA47F2, SD1, BA87D17, P3F8, BA43A11, SD4, SD5, SPUD1636, SD7 and GP179 are in coupling with GP21, and SD2, BA213C14, BA76O11, SD3 and SD6 are in repulsion with GP21. Bold horizontal lines represent chromosomal regions derived from the haplotype harbouring GP21 and thin horizontal lines represent chromosomal regions derived from the other haplotype. In the column on the right, the number of genotypes for each recombination event is given
High-resolution map of the Grp1 locus
The Grp1 locus harbours resistance to two potato cyst nematode species viz. a major resistance to Globodera pallida pathotype Pa2 and a major resistance to G. rostochiensis Ro5 (Rouppe van der Voort et al. 1998). In addition, a minor resistance to G. pallida Pa3 was detected (Rouppe van der Voort et al. 1998). It was shown that the QTLs for Pa3 coincided with the QTL for Pa2. Therefore, the 44 recombinants were tested for resistance to nematode population D383 (Pa2) and line 22 (Ro5).
Resistance was evaluated by counting the number of newly formed cysts on a subset of 54 progenies with recombination events between the markers GP21 and GP179. The progenies were split into two identical sets that were separately, but simultaneously, assessed for Ro5 and Pa2 resistances. The average number of cysts developed per plant genotype for each nematode population is presented in Suplementary Table 1. The resistant parent showed an average of 2 cysts per plant for Ro5 and 1 for Pa2, while on the susceptible parent 103 and 70 cysts per plant developed for Ro5 and Pa2, respectively.
Analysis of variance on normalised cyst counts revealed that the genetic variance for both G. pallida and G. rostochiensis resistance was significant (P < 0.0001). The log-transformed values of the averaged number of cysts per genotype showed a continuous distribution skewed towards susceptibility for both nematode populations. The broad-sense heritability was estimated to be 0.95 for G. rostochiensis and 0.93 for G. pallida. Interval mapping in the GP21–GP179 region was applied on the average number of cysts per genotype. For both G. pallida and G. rostochiensis, a significant QTL was detected with a maximum LOD score of 9.03 and 7.90, respectively. The QTL for G. pallida resistance explains 67% of total G. pallida resistance and the QTL for G. rostochiensis resistance explains 62% of total G. rostochiensis resistance. LOD score graphs for G. rostochiensis and G. pallida resistance are presented in Fig. 2.
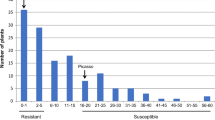
Grp1 resistance is located in an interval that is known to harbour single dominant R genes in other potato genotypes (Kuang et al. 2005; Ballvora et al. 2007). In addition, because PCN are obligatory outbreeding nematode species, the nematode populations used to test for resistance are most likely a mixture of virulent and avirulent genotypes (Janssen et al. 1990). Therefore, it is possible that the quantitative effect of the Grp1 resistance is caused by the genetic diversity of the nematodes and that resistance is monogenic. To test this, the plant genotypes were first sorted on the presence or absence of the closest marker, which is TG432 for both G. rostochiensis (LOD 7.3) and G. pallida (LOD 7.9) resistance, and then according to ascending cyst numbers (Fig. 3). For the majority of the genotypes, the presence of marker TG432 was indicative of the degree of resistance (Fig. 3). A small number of genotypes with an intermediate level of resistance lacked a correlation with marker TG432, indicating that also other regions in the potato genome affect the degree of resistance.
Distribution graphs for the number of G. rostochiensis cysts (a) and G. pallida cysts (b). The Y axes represent the number of cysts per plant. The X axes represent the potato genotypes. In the potato genotypes to the left of the dashed vertical line, marker TG432 is present and in the potato genotypes to the right of this line, marker TG432 is absent. Potato genotypes in the grey areas are classified as intermediate, while those above the grey areas are classified as susceptible and those below as resistant. Potato genotypes that are discussed in the text are indicated with a code
The G. pallida and G. rostochiensis resistance conferred by the Grp1 locus showed tight linkage in the test population of 1,536 potato genotypes. Both resistances could be mapped between markers SPUD839 (with nine recombination events) and TG432 (with one recombination event; Fig. 4). Eight susceptible recombinants clearly indicated that resistance conferred by Grp1 is located south of SPUD839 and one recombinant between marker TG432 and Grp1 (4A7) gives a clear resistant phenotype indicating that Grp1 resistance is located north of TG432. Two potato genotypes (3H1 and 1B12) have a recombination event between markers SPUD839 and TG432, but have intermediate levels of resistance for G. pallida and G. rostochiensis. It is, therefore, not clear where these recombination events take place exactly. Grp1 resistance behaves as a single gene; none of the recombinants between SPUD839 and TG432 showed a clear difference in resistance or susceptibility to either one of the two nematode species. Only the recombinant genotype 2H8 showed some, but inconclusive, difference in resistance level. This genotype was classified as susceptible for G. rostochiensis (90 cysts), but had an intermediate level of resistance for G. pallida (36 cysts; Fig. 3). One genotype (12C6) showed a marked difference in average cyst number to both PCN species (Fig. 3). However, this double recombination event results in a singleton. This genotype has a recombination event between group1 and marker SD2 (Fig. 1) and the four other genotypes that have a recombination event between group1 and SD2 show resistance and susceptibility as expected. Therefore, it is suspected that the G. rostochiensis resistance phenotype of genotype 12C6 is the result of a mistake, which is also corroborated by a large standard deviation for the cyst numbers of G. rostochiensis (Table 2), and this genotype was omitted from further analysis. Markers flanking the Grp1 locus define an interval of 1.08 cM. From these results, it can be concluded that PCN resistance conferred by the Grp1 locus may be attributed to a single gene or to two or more tightly linked genes.
Discussion
In this study, a high-resolution map of the broad-spectrum potato cyst nematode resistance locus Grp1 was constructed. Sixteen markers were identified within the 4 cM interval that harbours this locus. A recombination analysis followed by phenotyping for resistance resulted in the identification of significant QTLs for both G. rostochiensis and G. pallida resistance. The graphs of G. pallida and G. rostochiensis resistances are almost identical, both with the highest LOD score close to marker TG432. In this region, several other PCN resistance loci have been mapped. Gpa and GpaM1 in S. spegazzinii (Caromel et al. 2003; Kreike et al. 1994), GpaV s spl in S. sparsipilum (Caromel et al. 2005) and Gpa5 (Rouppe van der Voort et al. 2000). In addition another G. pallida resistance gene was also mapped in this region, which resembles the Gpa5 locus so much that it is suggested to be the same locus in S. vernei (Bryan et al. 2002; Rouppe van der Voort et al. 2000). All these loci confer resistance to G. pallida only and at least some of them have been introgressed in commercial potato varieties (Sattarzadeh et al. 2006). In contrast, Grp1 confers resistance to both G. pallida and G. rostochiensis (Rouppe van der Voort et al. 1998).
The Grp1 locus explains 62% of the total G. rostochiensis resistance and 68% of the total G. pallida resistance. This suggests that other places in the genome harbour minor resistance effects, which could be an explanation for the occurrence of the intermediate genotypes after the ordering based on the presence and absence of marker TG432 (Fig. 3). Remarkably, no significant QTLs have been detected elsewhere in the genome (Rouppe van der Voort et al. 1998). Interestingly, all G. pallida resistance loci that map in the GP21-GP179 interval on chromosome V are found in genotypes that harbour other QTLs for resistance. Kreike et al. (1994) and Caromel et al. (2003) found two other minor G. pallida resistance loci on chromosomes IV and VII, and VI and XII, respectively. For GpaV s spl and Gpa5 additive G. pallida resistance loci were identified on chromosomes XI and IX, respectively (Bryan et al. 2002; Caromel et al. 2005; Rouppe van der Voort et al. 2000).
Genome-wide sequence analysis and genetic mapping of R gene candidates have shown that R genes are often located in clusters spread throughout the plant genome (reviewed by Gebhardt and Valkonen 2001). Notably, QTLs conferring resistance to potato cyst nematodes often co-localise with clusters of R gene homologues. A Phythophthora infestans resistance locus, explaining 50% of the field resistance, resides in a region comprising R gene homologues of the NBS-LRR class (Tan et al. 2008). In addition, this resistance is associated with a hypersensitive response, normally resulting from dominant R genes of the NBS-LRR class. This indicates that R genes of the NBS-LRR class may contribute to partial resistance to P. infestans, a situation that may also apply to the quantitative resistance conferred by Grp1.
It is also noted that potato cyst nematodes reproduce by obligate outcrossing, and that PCN populations often consist of a mixture of virulent and avirulent genotypes (Bakker et al. 1993). As a consequence, quantitative resistance to PCN may be conferred by dominant R genes operating on the basis of a classical gene-for-gene relationship. Indeed, after dividing the recombinant genotypes used to fine map the Grp1 locus into classes of resistant, intermediate and susceptible genotypes for G. pallida and G. rostochiensis, both resistances behaved as single dominant R genes. The resistances to G. pallida and G. rostochiensis could not be separated by a recombination event suggesting that Grp1 resistance is conferred by a single R gene or tightly linked R genes. Preliminary comparative mapping (data not shown) indicates that Grp1 resistance is positioned in the same area as the homologues of R1, Bs4 and Prf. R1 and Bs4 mediate resistance to P. infestans in potato (Ballvora et al. 2002) and Xanthomonas campestris pv. vesicatoria in tomato (Schornack et al. 2004), respectively. Prf interacts with AvrPto (Salmeron et al. 1996). All three genes belong to the NBS-LRR class of resistance genes and could be used as candidate genes for Grp1.
Because so many G. pallida resistances have been mapped to the same region on chromosome V in various potato species, it can be suggested that several of these resistances have a common evolutionary background. This suggestion is strengthened by the fact that all these loci confer resistance to the pathotypes Pa2/3. In case of a common origin, the resistance specificity must be relatively old and arisen before the speciation of S. vernei, S. sparsipilum and S. spegazzinii. However, the resistance locus Grp1 confers not only resistance to G. pallida Pa2/3, but also to G. rostochiensis Ro5. The potato genotype that harbours Gpa5 is closely related to the genotype that harbours Grp1 and many common markers have been identified (Rouppe van der Voort et al. 2000). Because breeding was only focussed on G. pallida resistance, it was speculated that G. rostochiensis resistance was lost during selection (Rouppe van der Voort et al. 2000). Based on the comparison of common markers at the Gpa5 locus and the Grp1 locus, it was suggested that G. pallida resistance was conferred by a gene north of TG432 and G. rostochiensis resistance by a gene south of marker TG432. This is not confirmed by the high-resolution map of the Grp1 locus produced in this study, which suggests that both Pa2/3 and Ro5 resistances are conferred by a single gene, or two or more tightly linked genes just north of TG432.
If Grp1 resistance is conferred by a single gene, this gene has a dual specificity. So far, multiple specificities have been shown for Mi-1 and RPM1 (Grant et al. 1995; Nombela et al. 2003; Rossi et al. 1998; Vos et al. 1998). Mi-1 confers resistance to several species of root knot nematodes as well as to aphids and whiteflies (Milligan et al. 1998; Nombela et al. 2003; Rossi et al. 1998; Vos et al. 1998). It is unlikely that Mi-1 interacts with identical ligands from all these pests. Even within these pest species, variability in virulence occurs, indicating that ligand conservation among these taxonomically unrelated species is not very likely. Although G. pallida and G. rostochiensis are sibling species, it has been shown that they are extremely distinct at the molecular level (Bakker and Bouwman-Smits 1988). Like Mi-1 resistance, Grp1 resistance also shows within-species variability in virulence. Therefore, we assume that Grp1 does not recognise identical ligands from G. pallida and G. rostochiensis. It is more likely that Grp1, like RPM1 (Mackey et al. 2002) recognises indirectly (“guard model”) two unrelated effectors that modify the same host protein. Proof for such a dual specificity of Grp1 can only be given after the identification of the gene underlying this resistance locus, for which this study will form the basis.
References
Bakker J, Bouwman-Smits L (1988) Contrasting rates of protein and morphological evolution in cyst nematode species. Phytopathology 78:900–904
Bakker J, Folkertsma RT, Rouppe van der Voort JNAM, De Boer JM, Gommers FJ (1993) Changing concepts and molecular approaches in the management of virulence genes in potato cyst nematodes. Ann Rev Phytopathol 31:169–190
Ballvora A, Ercolano MR, Weiss J, Meksem K, Bormann CA, Oberhagemann P, Salamini F, Gebhardt C (2002) The R1 gene for potato resistance to late blight (Phytophthora infestans) belongs to the leucine zipper/NBS/LRR class of plant resistance genes. Plant J 30:361–371
Ballvora A, Jocker A, Viehover P, Ishihara H, Paal J, Meksem K, Bruggmann R, Schoof H, Weisshaar B, Gebhardt C (2007) Comparative sequence analysis of Solanum and Arabidopsis in a hotspot for pathogen resistance on potato chromosome V reveals a patchwork of conserved and rapidly evolving genome segments. Bmc Genomics 8:112
Barone A, Ritter E, Schachtschabel U, Debener T, Salamini F, Gebhardt C (1990) Localization by restriction-rragment-length-polymorphism mapping in potato of a major dominant gene conferring resistance to the potato cyst nematode Globodera rostochiensis. Mol Gen Genet 224:177–182
Bendahmane A, Querci M, Kanyuka K, Baulcombe DC (2000) Agrobacterium transient expression system as a tool for the isolation of disease-resistance genes: application to the Rx2 locus in potato. Plant J 21:73–81
Bonierbale MW, Plaisted RL, Pineda O, Tanksley SD (1994) QTL analysis of trichome-mediated insect resistance in potato. Theor Appl Genet 87:973–987
Bryan GJ, McLean K, Bradshaw JE, De Jong WS, Phillips M, Castelli L, Waugh R (2002) Mapping QTLs for resistance to the cyst nematode Globodera pallida derived from the wild potato species Solanum vernei. Theor Appl Genet 105:68–77
Caromel B, Mugniéry D, Lefebvre V, Andrzejewski S, Ellissèche D, Kerlan MC, Rousselle P, Rousselle-Bourgeois F (2003) Mapping QTLs for resistance against Globodera pallida (Stone) Pa2/3 in a diploid potato progeny originating from Solanum spegazzinii. Theor Appl Genet 106:1517–1523
Caromel B, Mugniery D, Kerlan MC, Andrzejewski S, Palloix A, Ellisseche D, Rousselle-Bourgeois F, Lefebvre V (2005) Resistance quantitative trait loci originating from Solanum sparsipilum act independently on the sex ratio of Globodera pallida and together for developing a necrotic reaction. Mol Plant Microbe Interact 18:1186–1194
De Jong W, Forsyth A, Leister D, Gebhardt C, Baulcombe D (1997) A potato hypersensitive resistance gene against potato virus X maps to a resistance gene cluster on chromosome 5. Theor Appl Genet 95:246–252
Dellaert LMW, Vinke JH (1987) Testing potatoes for resistance to Globodera pallida pathotype Pa-3: resistance spectra of plant genotypes and virulence spectra of Pa-3 isolates. Rev Nematol 10:445–453
Gebhardt C, Valkonen JPT (2001) Organization of genes controlling disease resistance in the potato genome. Ann Rev Phytopathol 39:79–102
Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL (1995) Structure of the Arabidopsis Rpm1 gene enabling dual-specificity disease resistance. Science 269:843–846
Janssen R, Bakker J, Gommers FJ (1990) Assessing intra-specific variations in virulence in Globodera rostochiensis and Globodera pallida Rev. Nematology 13:11–16
Kreike CM, Dekoning JRA, Vinke JH, Vanooijen JW, Stiekema WJ (1994) Quantitatively inherited resistance to Globodera pallida is dominated by one major locus in Solanum spegazzinii. Theor Appl Genet 88:764–769
Kuang HH, Wei FS, Marano MR, Wirtz U, Wang XX, Liu J, Shum WP, Zaborsky J, Tallon LJ, Rensink W, Lobst S, Zhang PF, Tornqvist CE, Tek A, Bamberg J, Helgeson J, Fry W, You F, Luo MC, Jiang JM, Buell CR, Baker B (2005) The R1 resistance gene cluster contains three groups of independently evolving, type I R1 homologues and shows substantial structural variation among haplotypes of Solanum demissum. Plant J 44:37–51
Lander ES, Botstein D (1989) Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121:185–199
Leonards-Schippers C, Gieffers W, Salamini F, Gebhardt C (1992) The R1 gene conferring race-specific resistance to Phytophthora infestans in potato is located on potato chromosome V. Mol Gen Genet 233:278–283
Leonards-Schippers C, Gieffers W, Schaefer PR, Ritter E, Knapp SJ, Salamini F, Gebhardt C (1994) Quantitative resistance to Phytophthora infestans in potato: a case study for QTL mapping in an allogamous plant species. Genetics 137:67–77
Mackey D, Holt BF, Wiig A, Dangl JL (2002) RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108:743–754
Milligan SB, Bodeau J, Yaghoobi J, Kaloshian I, Zabel P, Williamson V (1998) The root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell 10:1307–1319
Nombela G, Williamson VM, Muniz M (2003) The root-knot nematode resistance gene Mi-1.2 of tomato is responsible for resistance against the whitefly Bemisia tabaci. Mol Plant Microbe Interact 16:645–649
Oerke EC, Dehne HW, Schonbeck F, Weber A (1994) Crop production and crop protection: estimated losses in major food and cash crops. Elsevier Science B.V, Amsterdam
Paal J, Henselewski H, Muth J, Meksem K, Menendez CM, Salamini F, Ballvora A, Gebhardt C (2004) Molecular cloning of the potato Gro1–4 gene conferring resistance to pathotype Ro1 of the root cyst nematode Globodera rostochiensis, based on a candidate gene approach. Plant J 38:285–297
Park TH, Vleeshouwers V, Huigen DJ, van der Vossen EAG, van Eck HJ, Visser RGF (2005) Characterization and high-resolution mapping of a late blight resistance locus similar to R2 in potato. Theor Appl Genet 111:591–597
Riano-Pachon DM, Nagel A, Neigenfind J, Wagner R, Basekow R, Weber E, Mueller-Roeber B, Diehl S, Kersten B (2009) GabiPD: the GABI primary database, a plant integrative ‘omics’ database. Nucl Acids Res 37:D954–D959
Ritter E, Debener T, Barone A, Salamini F, Gebhardt C (1991) RFLP mapping on potato chromosomes of 2 genes controlling extreme resistance to potato virus X (PVX). Mol Gen Genet 227:81–85
Rossi M, Goggin FL, Milligan SB, Kaloshian I, Ullman DE, Williamson VM (1998) The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc Natl Acad Sci USA 95:9750–9754
Rouppe van der Voort J, Wolters P, Folkertsma R, Hutten R, Van Zandvoort P, Vinke H, Kanyuka K, Bendahmane A, Jacobsen E, Janssen R, Bakker J (1997) Mapping of the cyst nematode resistance locus Gpa2 in potato using a strategy based on comigrating AFLP markers. Theor Appl Genet 95:874–880
Rouppe van der Voort JR, Lindeman W, Folkertsma R, Hutten R, Overmars H, van der Vossen E, Jacobsen E, Bakker J (1998) A QTL for broad-spectrum resistance to cyst nematode species (Globodera spp.) maps to a resistance gene cluster in potato. Theor Appl Genet 96:654–661
Rouppe van der Voort J, Van der Vossen E, Bakker E, Overmars H, Van Zandvoort P, Hutten R, Klein-Lankhorst R, Bakker J (2000) Two additive QTLs conferring broad-spectrum resistance in potato to Globodera pallida are localized on resistance gene clusters. Theor Appl Genet 101:1122–1130
Salmeron JM, Oldroyd GED, Rommens CMT, Scofield S, Kim H-S, Lavelle DT, Dahlbeck D, Staskawicz B (1996) Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the pto kinase gene cluster. Cell 86:123–133
Sattarzadeh A, Achenbach U, Lubeck J, Strahwald J, Tacke E, Hofferbert HR, Rothsteyn T, Gebhardt C (2006) Single nucleotide polymorphism (SNP) genotyping as basis for developing a PCR-based marker highly diagnostic for potato varieties with high resistance to Globodera pallida pathotype Pa2/3. Mol Breed 18:301–312
Schornack S, Ballvora A, Gurlebeck D, Peart J, Baulcombe D, Ganal M, Baker B, Bonas U, Lahaye T (2004) The tomato resistance protein BS4 is a predicted non-nuclear TIR-NB-LRR protein that mediates defense responses to severly truncated derivatives of AvrBs4 and overexpressed AvrBs3. Plant J 37:787
Tan MYA, Hutten RCB, Celis C, Park T-H, Niks RE, Visser RGF, van Eck HJ (2008) The RPi-mcd1 locus from Solanum microdontum involved in resistance to Phytophthora infestans, causing a delay in infection, maps on potato chromosome 4 in a cluster of NBS-LRR genes. Mol Plant Microbe Interact 21:909–918
Van der Vossen E, Rouppe van der Voort J, Kanyuka K, Bendahmane A, Sandbrink H, Baulcombe D, Bakker J, Stiekema W, Klein-Lankhorst R (2000) Homologues of a single resistance-gene cluster in potato confer resistance to distinct pathogens: a virus and a nematode. Plant J 23:567–576
Van Ooijen JW (2004) MapQTL (R) 5, software for the mapping of quantitative trait loci in experimental populations. Kyazma BV, Wageningen
Vos P, Simons G, Jesse T, Wijbrandi J, Heinen L, Hogers R, Frijters A, Groenendijk J, Diergaarde P, Reijans M, Fierens-Osterenk J, De Both M, Peleman J, Liharska T, Hontelez J, Zabeau M (1998) The tomato Mi-1 gene confers resistance to both root-knot nematodes and potato aphids. Nature Biotech 16:1365–1369
Acknowledgments
This research was financially supported by the Dutch Technology Foundation (STW) and EC grant FAIR5-PL97-2565. We thank the potato breeding company HZPC Holland B.V. Research & Development (Metslawier) for the production of tubers from the in vitro population of recombinants, which enabled us to perform greenhouse nematode resistance assays, and Dieuwertje Lont and Kamila Koropacka for their assistance with the nematode resistance assays.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. Gebhardt.
A. Finkers-Tomczak and S. Danan contributed equally to this research.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Finkers-Tomczak, A., Danan, S., van Dijk, T. et al. A high-resolution map of the Grp1 locus on chromosome V of potato harbouring broad-spectrum resistance to the cyst nematode species Globodera pallida and Globodera rostochiensis . Theor Appl Genet 119, 165–173 (2009). https://doi.org/10.1007/s00122-009-1026-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-009-1026-1