Abstract
Cowpea is an important crop for subsistence farmers in arid regions of Africa, Asia, and South America. Efforts to develop cultivars with improved productivity under drought conditions are constrained by lack of molecular markers associated with drought tolerance. Here, we report the mapping of 12 quantitative trait loci (QTL) associated with seedling drought tolerance and maturity in a cowpea recombinant inbred (RIL) population. One hundred and twenty-seven F8 RILs developed from a cross between IT93K503-1 and CB46 were screened with 62 EcoR1 and Mse1 primer combinations to generate 306 amplified fragment length polymorphisms for use in genetic linkage mapping. The same population was phenotyped for maintenance of stem greenness (stg) and recovery dry weight (rdw) after drought stress in six greenhouse experiments. In field experiments conducted over 3 years, visual ratings and dry weights were used to phenotype drought stress-induced premature senescence in the RIL population. Kruskall–Wallis and multiple-QTL model mapping analysis were used to identify QTL associated with drought response phenotypes. Observed QTL were highly reproducible between stg and rdw under greenhouse conditions. Field studies confirmed all ten drought-response QTL observed under greenhouse conditions. Regions harboring drought-related QTL were observed on linkage groups 1, 2, 3, 5, 6, 7, 9, and 10 accounting for between 4.7 and 24.2% of the phenotypic variance (R 2). Further, two QTL for maturity (R 2 = 14.4–28.9% and R 2 = 11.7–25.2%) mapped on linkage groups 7 and 8 separately from drought-related QTL. These results provide a platform for identification of genetic determinants of seedling drought tolerance in cowpea.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cowpea [Vigna unguiculata. (L.) Walp.] is a crop of major economic importance in the arid and semi-arid regions of Africa, Asia, Latin America and to a lesser extent southeastern United States (Ehlers and Hall 1997). Recent estimates indicate that over 12.5 million tons of cowpea grain are produced worldwide, with Africa’s arid Sahel region accounting for 64% of total cowpea seed production (Fery 2002). In addition to its high nutritional value and ability to fix nitrogen in relatively poor soil, cowpea has greater drought tolerance than most other crop plants (Ehlers and Hall 1997; Hall 2004a). Hence, cowpea is comparatively productive in areas of marginal soil fertility and low rainfall (Sanginga et al. 2000).
In the Sudanian and Sahelian semi-arid regions where cowpea is widely cultivated, the frequency and intensity of droughts have increased over the last 30 years (Hall et al. 2003) due to climatic changes and human activities (Wittig et al. 2007), causing a shift to more drought tolerant crops such as cowpea (Van Duivenbooden et al. 2002). Given that these trends will likely continue there is increasing need to identify and develop drought tolerant crop cultivars for these agro-ecological zones.
Despite its inherent drought tolerance, significant differences in response to drought stress exist among cowpea genotypes (Hall et al. 2003; Hall 2004b; Muchero et al. 2008). However, an understanding of the genetic basis of drought tolerance in cowpea is lacking because few studies have been carried out to isolate drought responsive genetic elements or identify regions of the cowpea genome involved in drought stress response. Several gene transcripts have been isolated in response to drought-induced desiccation of cowpea. cDNAs homologous to 12-oxo-phytodienoic acid reductase, NADPH-dependent aldehyde reductase, alcohol dehydrogenase, dehydrin (Iuchi et al. 1996a), lipoxygenase (Iuchi et al. 1996b), multicystanin (Diop et al. 2004), 9-cis-epoxycarotenoid dioxygenase (VuNCED1) (Iuchi et al. 2000), phospholipase D (El-Maarouf et al. 1999), galactolipid acyl hydrolase (Matos et al. 2001), phosphatidylinositol-specific phospholipase C (El-Maarouf et al. 2001), ascorbate peroxidase (D’Arcy-Lameta et al. 2006) and more recently glutathione reductase (Contour-Ansel et al. 2006) have been described. However, their individual comparative contributions toward cowpea drought tolerance have not been investigated. Of these cDNAs, VuNCED1 was tested in transgenic bent grass where its stable expression led to a marked increase in drought tolerance (Aswath et al. 2005). The nature of these isolated transcripts and their pathways is highly suggestive of senescence-based drought tolerance mechanisms. Previously, we described phenotypic responses observed among cowpea genotypes exposed to drought stress during the early vegetative stage (Muchero et al. 2008). Notably, maintenance of stem greenness was highly correlated with survival and recovery after drought stress under both greenhouse and field conditions.
In this study, a recombinant inbred population derived from seedling drought tolerant line IT93K503-1 and susceptible CB46 (Muchero et al. 2008) was used to identify quantitative trait loci (QTL) within the cowpea genome that are significantly associated with seedling and young plant survival during severe drought stress. In addition, an evaluation was made of the genetic overlap between regions associated with drought stress-induced premature senescence (from here forward referred to as ‘drought-induced senescence’) and maturity. Maturity is known to confound studies of senescence-based drought tolerance in cowpea (Watanabe et al. 1997). Therefore, we attempted to distinguish between QTL mediating drought-induced senescence versus senescence resulting from plant maturity. This paper reports on cowpea QTL that were found to be significantly associated with drought tolerance as measured by recovery dry weight, stem greenness, and field drought-induced senescence. Separate QTL associated with maturity are also described.
Materials and methods
Plant materials
One hundred and twenty-seven F8 recombinant inbred lines (RILs) developed by single seed descent from a cross between seedling drought tolerant IT93K503-1 and susceptible CB46 homozygous cowpea genotypes were used as the mapping population. IT93K503-1 is a medium-maturing variety developed by the International Institute of Tropical Agriculture (IITA) while CB46 is an early-maturing variety developed for irrigated production in California, USA. An initial subset of 57 RILs was advanced to F8 and was used in greenhouse experiments 1-4 and field experiments CVARS 2005 and CVARS 2006. The remaining 70 lines were advanced to F8 separately and used together with the initial 57 lines in greenhouse experiments 5 and 6 as well as field experiment CVARS 2007. All seeds used for experiments and molecular studies were grown and harvested under uniform greenhouse conditions.
Greenhouse experiments 1 and 2
All greenhouse experiments were carried out at the University of California Riverside campus (33°57′54″N; 117°20′08″W). In March 2005, two side-by-side experiments were carried out in 878-ml and 2-l pots filled with UCMIX-B (Matkin and Chandler 1957). Four replicates of 57 RILs and the two parental lines were arranged in a randomized complete block design under greenhouse conditions with temperature set at 22°C at night and 32°C during the day. Day lengths for the duration of the experiments were approximately 11 h 40 min. Small pots (878 ml) were planted with two seeds and later thinned to one plant per pot 9 days after planting. Large pots (2 l) were planted with four seeds and later thinned to two plants per pot 9 days after planting. Irrigation was maintained until thinning when water was withheld for 28 days in 878-ml pots and for 40 days in 2-l pots. Visual scores for stem greenness (0–5 scale, 0 = yellow, 5 = deep green, see Muchero et al. 2008) were taken on days 10, 12, 14, 16, 18, 20, 22, 24, 26, and 28 after last watering. Three weeks were allowed for recovery after re-introduction of irrigation. Surviving plants were harvested for recovery dry weight determination. Prematurely senesced tissue was trimmed leaving only surviving green tissue before drying in an oven at 65°C until no change in weight was observed.
Greenhouse experiments 3 and 4
Two separate experiments were conducted in March and May 2006 with 57 RILs and the two parental lines using 878-ml pots. Each experiment had four replicates arranged in a randomized complete block design. Greenhouse temperature was set between 32 and 35°C during the day and 22 and 25°C at night. Day lengths were approximately 12 h during March and 14 h in May. Individual pots were planted with two seeds and were thinned to one plant per pot 8 days after germination. After thinning, pots were watered to capacity after which water was withheld for 27 days in experiment 3 and for 34 days in experiment 4. Scores were taken for stem greenness as described above. Three weeks were allowed for recovery after watering was resumed before surviving plants were harvested for recovery dry weight determination as described for experiments 1 and 2.
Greenhouse experiments 5 and 6
Two separate experiments were conducted using 127 RILs (Greenhouse 5) and 125 RILs (Greenhouse 6), respectively. In both experiments 878-ml pots filled with UCMIX-B were used. Greenhouse experiment 5 was planted on January 24, 2008 and Greenhouse experiment 6 on March 6, 2008. The same experimental and phenotyping protocols as described for greenhouse experiments 1–4 were followed for both experiments. Temperature settings were kept at 25–27°C at night and 32–35°C during the day for both experiments. Light and darkness cycles were not artificially controlled and day lengths were around 11 h during experiment 5 and 12 h during experiment 6.
Field experiments for drought
Field experiments CVARS 2005, CVARS 2006, and CVARS 2007 were carried out at the University of California-Riverside Coachella Valley Agricultural Research Station (CVARS) (33°37′52″N, 116°06′43″W). The experiments were planted on August 7, 2005 (CVARS 2005); August 9, 2006 (CVARS 2006); and June 19, 2007 (CVARS 2007). The experimental set-up was the same for all three experiments except 57 RILs and the two parental lines were used in experiments CVARS 2005 and CVARS 2006 whereas 124 RILs and the two parents were planted in experiment CVARS 2007. Label rate applications of Treflan or Treflan–Dual preplant herbicide were made to control weeds. Four seeds were planted every 30-cm in 5-m-long plots on rows 75 cm apart with a two-row cone planter. Each genotype was replicated four times in a randomized complete block design. All experiments were brought to field capacity using drip-irrigation immediately after planting. Plots for experiments CVARS 2005 and CVARS 2006 received 1 h of sprinkler-irrigation after planting to incorporate Treflan herbicide. No additional irrigation was applied for the duration of the experiments. Visual scores for drought-induced senescence were taken every 14 days for the duration of the experiment on a 0–10 scale (0–1 = 0 to <5% senescence, 2–3 = 5–10% damage, 3–4 = 10–15% damage, 5–6 = 15–25%, 7–8 = 25–50%, 9–10 = >50% senescence). At the end of each experiment, five plants per plot were sampled at random for dry weight-based determination of percent leaf damage. Ten randomly selected leaves were taken for drought-induced senescence determination. Senesced portions of the leaves were trimmed and weighed to give dry weight of senesced leaf tissue for each replicate. The non-senesced portions were oven-dried until no change in weight was observed, to give the dry weight of non-senesced leaf tissue. Percent damage for each replicate was calculated using the equation: % leaf damage = [dry weight of senesced tissue/(dry weight of senesced tissue + dry weight of non-senesced tissue)]100. Experimental periods, average temperatures, evapo-transpiration, and precipitation recorded during the experiments are given in Table 1.
Field experiments for maturity
Experimental plots for all genotypes were set up as described for the field-based drought experiments. 57 RILs and two parental lines were evaluated in experiments at Riverside in 2006, Coachella in 2006, and Riverside in 2007. Also, in the experiment at Riverside in 2008 108 RILs and the two parental lines were evaluated for maturity. Location, experimental period, temperature, and evapo-transpiration conditions during maturity field experiments are described in Table 1. The experiments were planted on May 16, 2006 (Riverside 2006); August 9, 2006 (Coachella 2006); May 22, 2007 (Riverside 2007); and June 17, 2008 (Riverside 2008). All plots were furrow-irrigated once per week at Riverside and drip-irrigated twice per week at the Coachella Valley station. Scores for maturity were taken every 14 days beginning at the time when the earliest maturing parental genotype (CB46) was fully podded. Maturity was rated on a 0–10 scale based on leaf senescence as described for drought-induced senescence.
AFLP analysis
Genomic DNA was isolated from young fresh trifoliates of greenhouse-grown cowpea plants using Qiagen mini kit as per manufacturer’s protocol. DNA was quantified using a TD-360 Mini-fluorometer (Turner Designs, Sunnyville, CA, USA). Working DNA solutions were obtained by dilution with ddH2O to 10 ng/μl. AFLP genotyping (Vos et al. 1995) was carried out according to the specification for IRDye™ Fluorescent AFLP® Kit for Large Plant Genome Analysis with minor modifications as reported by Fang et al. (2005). Briefly, 500 ng in 9 μl genomic DNA was incubated for 2 h at 37°C and then 70°C for 15 min in 2.5 μl manufacturer-supplied 5× reaction buffer and 1.0 μl EcoR1/Mse1 enzyme mix. Adaptors were ligated by adding 12 μl adaptor mix and 0.5 μl T4 DNA ligase. The mixture was incubated at 20°C for 2 h. A 1:5 dilution of the ligation mixture was made using TE buffer before the next step. Pre-amplification of the diluted ligation mixture was carried out with the following reaction mixture: 2.5 μl diluted DNA in 20.0 μl AFLP Pre-amp primer mix, 2.5 μl 10× PCR reaction buffer, 0.5 μl Taq DNA polymerase (5 U/μl). PCR reactions were carried out in a Mastercycler® gradient thermocycler (Eppendorf AG, Hamburg, Germany) with the following program; 30 cycles at 94°C for 30 s, 56°C for 1 min, and 72°C for 1 min. A 1:25 dilution was made using ddH2O after the reaction. Samples were stored at −20°C until further use. Selective amplification was done with 62 primer combinations. Each reaction duplex was carried out in 13 μl reaction volume containing 3 μl DNA sample, 1.42 μl three-selective base Mse1 primer, 0.75 μl each of two three-selective base-IRDye-labeled EcoR1 (IRDye 700 and IRDye 800) primers (LI-COR Biotechnology, Lincoln, NE, USA), 4.73 μl ddH2O, 1.42 μl 10× PCR reaction buffer, 0.7 μl MgCl2 (50 mM) and 0.23 μl Taq DNA polymerase (5 U/μl). Cycling parameters were as follows: 1 cycle of 2 min at 94°C, 30 s at 65°C, and 1 min at 72°C. This was followed by 10 cycles in which a step-wise reduction by 1°C of annealing temperature was done with other steps unchanged. This was followed by 29 cycles of 30 s at 94°C, 30 s at 56°C, and 1 min at 72°C. PCR product was mixed with 12.5 μl blue stop solution (9,800 μl formamide, 200 μl 0.5 M EDTA and 0.01 g bromophenol blue). The PCR product and stop solution mixture was stored at −20°C before use.
Samples were denatured at 95°C for 3 min and immediately returned to ice before electrophoresis. 0.8 μl samples were loaded onto 8% denaturing polyacrylamide [40 ml Long Ranger (Cambrex Bioscience Rockland Inc., ME, USA), 105 g urea, 20 ml 10× TBE) and separated in 0.8× TBE buffer using the LI-COR Automated Sequencer 4000-L (LI-COR Biotechnology, Lincoln, NE, USA). Gel apparatus was prerun at 1,500 V, 40 mA and 40 W for 45 min before samples were loaded. Separation of denatured products was carried out at 1,500 V for 3 h 10 min. Gel images were scored manually with polymorphic band presence noted as “1” and absence as “0”. Multiple polymorphic fragments generated by the same primer combination were numbered according to decreasing size order and were prefixed by six letter identification of the primer combination. The EcoR1 primer three-selective base sequences preceded the Mse1 three-selective base sequence in the naming nomenclature.
Map construction
Joinmap 3.0 (Van Ooijen and Voorrips 2001) was used to construct the genetic linkage map. The Kosambi mapping function (Kosambi 1944) was used to convert recombination frequencies to Centimorgans. A step-wise reduction of the LOD score from 7 to 3 with a maximum recombination of 45% was used to assign markers to linkage groups. Only markers showing the highest congruency were used in map construction.
QTL analysis
MapQTL 4.0 software (Van Ooijen et al. 2002) was used for QTL analysis. First, the non-parametric Kruskall–Wallis package was used to identify markers associated with drought and maturity response phenotypes. Data for each experiment and for each trait were analyzed separately. In order to analyze the effect of population size on QTL detection, data for drought greenhouse experiments 5, 6, and CVARS 2007, were mapped using subpopulations of 57 and 70 first and finally for the full ≥124 RIL population. For maturity, experiment Riverside 2008, subpopulations of 57 and 51 were used to map QTL separately for comparison with QTL detected using the 108 RIL population. Markers showing significant association at the more stringent 0.005 significance level (suggested by the authors of the software, Van Ooijen et al. 2002) or higher in at least one experiment were considered putative. Putative QTL were confirmed by the multiple-QTL model mapping (MQM) package using the automatic cofactor selection option (Visker et al. 2003; Curley et al. 2005). Given the large difference in linkage group sizes, LOD thresholds were determined at the 0.05 significance level for each linkage group and each experiment using the permutation test (Doerge and Rebai 1996). One thousand permutations were performed during each analysis. The 50th highest LOD was taken as the LOD significance threshold for each linkage group and each experiment, then the highest LOD significance threshold for each linkage group across all experiments was used as the final LOD threshold to identify QTL in all experiments. QTL were considered significant when the significance thresholds were met concurrently in at least two experiments for both the Kruskall–Wallis and the MQM analysis. QTL were considered distinct and separate when a drop in LOD scores of 1 was observed between respective peaks of the QTL over multiple experiments (Posthuma et al. 2005). Graphical representation of linkage groups and QTL was carried out using MapChart 2.2 software (Voorrips 2002).
Statistical analysis
Spearman rank correlation and frequency distribution analysis of the phenotypic data were carried out using the Statistix 8 package (Analytical Software 2003). Standard errors were calculated using LOD scores and R 2 estimates based on drought experiments reported in Supplementary materials for each respective QTL using the Statistix software. Standard errors for maturity QTL were calculated similarly using LOD scores and R 2 estimates from all four maturity experiments.
Results
AFLP analysis and linkage mapping
Of the 62 EcoR1 and Mse1 primer pairs screened on the population of 127 RILs, 8 primer combinations yielded no polymorphic markers. The remaining 54 primer pairs yielded 378 polymorphic markers. Of these, 20 markers exhibited significant segregation distortion at the 0.05 significance level as calculated by the JoinMap program, 22 markers were polymorphic in the RIL population but were present in both parents, and a further 12 markers were absent in both parents but were segregating in the RIL population. The latter 34 markers were not used in linkage mapping attempts but were analyzed in QTL analysis as unmapped markers. Three hundred and six AFLP markers were successfully assigned to 11 linkage groups (numbered LGI–LG11) spanning a total genetic distance of 643 cM (Fig. 1).
Location of drought-induced senescence and maturity QTL on a cowpea genetic linkage map constructed using 306 amplified fragment length polymorphism (AFLP) markers segregating in 127-F8 cowpea recombinant inbred lines developed from a cross between homozygous genotypes IT93K503–1 and CB46. Drought-induced senescence QTL are represented with solid bars and maturity QTL are represented by hatched bars
Drought-induced senescence and maturity
There was high correlation between visual stem greenness (stg) scores and recovery dry weight (rdw) within individual greenhouse experiments (0.6375 ≤ r ≤ 0.9369) (Table 2). Further, with the exception of greenhouse experiment 5, stg and rdw traits were significantly correlated across greenhouse experiments (r ≥ 0.3985) (Table 2). In general, field drought-induced senescence and greenhouse stg and rdw traits were negatively correlated. Significant correlations at the 0.05 probability level ranged from r = −0.2296 to −0.5415 between these traits (Table 2). Correlations between individual drought field experiments ranged from r = 0.1716 to 0.5409 (Table 2). Maturity field experiments were all highly correlated (r ≥ 0.7760, p = 0.000).
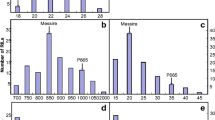
Cowpea genotypes IT93K503-1 and CB46 exhibited significant differences in tolerance to seedling-stage drought stress (Fig. 2a–c). IT93K503-1 displayed strong ability to maintain a viable green stem and survive greenhouse drought stress up to 4 weeks after last watering (Muchero et al. 2008). IT93K503-1 also exhibited minor drought-induced senescence under field drought conditions with only slight tip burning observed on the leaves. Compared to IT93K503-1, CB46 exhibited reduced ability to maintain a green stem and survive in response to greenhouse drought stress and displayed elevated drought-induced leaf senescence under field conditions.
These parental phenotypes also segregated in the RIL population. Frequency distribution of the phenotypic data deviated from normality and suggested multigene activity with transgressive segregants in both susceptible and tolerant phenotypes (Fig. 2a–c). For maturity, CB46 displayed maturity-related senescence under well-watered conditions earlier than IT93K503-1. Generally, a maturity difference of 25-30 days was observed between the two genotypes. Phenotypic frequency distribution for this trait was skewed toward the early maturity phenotype of CB46 (Fig. 2d). Transgressive segregants were also observed for maturity with some RILs showing earlier and others showing later maturity than parental extremes.
QTL analysis
Ten QTL were identified for seedling drought-induced senescence using recovery dry weight, visual stem greenness, visual drought-induced senescence, and percent leaf damage under greenhouse and field conditions (Table 3; Supplementary material). Both Kruskall–Wallis and MQM analysis identified the same QTL intervals (Table 3; Supplementary material). Due to the high correlation between visual stem greenness ratings and recovery dry weight (Table 2), there was high agreement between QTL identified using both assays under greenhouse conditions. This was true for all QTL, and examples of this relationship are given in Fig. 3, for Dro-1 and Dro-5. Similarly, visual drought-induced senescence and percent leaf damage as measured by dry weight in field experiments revealed the same QTL intervals (Fig. 4). Consistent results were obtained between greenhouse experiments, between greenhouse and field experiments, as well as between individual field experiments. All ten QTL were identified under field conditions in at least one experiment (Supplementary material). QTL Dro-3 on linkage group 2 was detected with high significance in three field experiments with R 2 estimates of 19.3, 22.8, and 8.8% (Supplementary material). However, there was indication that the population of 57 RILs resulted in inflated R 2 estimates for this QTL (Fig. 5). Dro-7 QTL on linkage group 6 was also detected under all three field experiments although at lower significance level. QTL Dro-1, Dro-2, Dro-8, and Dro-10 were detected at least twice under field conditions. Of these, QTL Dro-8 was the most stably expressed based on R 2 estimates of 12.6% in CVARS 2006 (57 RIL) and 12.8% in CVARS 2007 (124 RIL). The different population sizes did not affect R 2 estimates of this QTL, however statistical significance increased markedly with the larger population size. The largest proportion of phenotypic variance under field conditions was explained by QTL Dro-4 with R 2 = 19.6% in experiment CVARS 2007 conducted with 124 RIL (Supplementary material). This was followed by Dro-10 with R 2 = 15.2% and Dro-8 with R 2 = 12.8%. Dro-6 had the lowest detectability among all drought-induced senescence QTL. The QTL achieved statistical significance in only one experiment but was consistently detected in three greenhouse and one field experiments.
LOD traces for QTL a Dro–1 on linkage group 1 based on recovery dry weight (dashed line) and visual senescence ratings (solid line), b Dro-5 on linkage group 5 based on recovery dry weight (dashed line) and visual senescence ratings (solid line) for 127 recombinant inbred lines subjected to greenhouse drought stress. Horizontal broken line represents LOD significance threshold estimated from 1,000 permutations at the 0.05 probability level
LOD traces for drought-induced senescence QTL Dro-8 on linkage group 7 observed in greenhouse experiment 5 and field experiment CVARS 2007 (solid lines) and maturity QTL Mat-1 observed under field conditions in experiments Riverside 2006 and Coachella 2006 (dashed lines). Broken horizontal line represents LOD significance threshold estimated from 1,000 permutations at the 0.05 probability level
Effect of mapping population size on a LOD scores and b percent phenotypic variance explained (R 2) for drought-induced senescence QTL Dro-1 to Dro-10 and maturity QTL Mat-1 and Mat-2 in a cowpea recombinant inbred population (RIL) developed from a cross between IT93K503-1 and CB46. Data were based on QTL mapping analysis conducted with 57, 70, and ≥124 RIL for Dro-1 to Dro-10 and 57, 51, and 108 RIL for Mat-1 and Mat-2. Bars indicate ±SE. Dotted bar 57 RIL; broken striped bar 70 RIL (51 RIL for Mat-1 and Mat-2); striped bar ≥124 RIL (108 RIL for Mat-1 and Mat-2)
Quantitative trait loci for maturity were highly reproducible across the four experiments and were located on linkage groups 7 and 8 (Fig. 1). Mat-1 and Mat-2 QTL were consistently identified in the same interval in all experiments. Mat-1 explained 16.8-27.9% of the phenotypic variance in experiments conducted with 57 RIL, however, the phenotypic variance explained was slightly lower, 14.4%, when 108 RIL were evaluated. Similarly, Mat-2 explained 19.2–25.2% of the phenotypic variance in experiments using 57 RIL, but explained 11.7% of the phenotypic variance when 108 RIL were evaluated (Table 4). However, the location of both QTL remained stable regardless of the number of RIL evaluated (Table 4). The maturity QTL Mat-1 on linkage group 7 mapped relatively close to drought-induced senescence QTL Dro-8, although there was no apparent overlap between the two (Fig. 4).
The LOD significance thresholds differed substantially between larger and smaller linkage groups (Supplementary materials). For example, the highest LOD significance threshold based on 1,000 permutations was 2.1 for LG 3 and the lowest was 1.4 for LG 10. However, these thresholds did not differ significantly between different experiments for each respective linkage group. Differences in LOD significance thresholds between experiments were typically smaller or equal to 0.1 for each linkage group (data not shown).
Effect of population size on QTL detectability
Results of QTL mapping using the 57 and 70 RIL sub-populations were consistent with mapping results from the ≥124 RIL populations (Fig. 5). In general, the larger populations resulted in higher LOD scores for QTL identified (Fig. 5a). However, the 70 RIL sub-population revealed lower LOD scores for QTL Dro-3 compared to the 57 RIL sub-population, whereas the 57 and the ≥124 RIL populations showed no difference for the same QTL (Fig. 5a). Compared to the larger ≥124 RIL populations, the 57 RIL sub-populations mapped the same relative QTL interval for nine out of the ten QTL identified in separate greenhouse and field experiments (Supplementary material). The only exception was QTL Dro-5, which mapped to opposite ends of the same linkage group depending on the population size (Supplementary material). In general, the smaller population resulted in higher estimates for phenotypic variance (R 2) explained by individual QTL (Fig. 5b). Differences between R 2 estimates based on the 57 and 70 RIL sub-populations compared to the ≥124 RIL populations were statistically significant for QTL Dro-2, Dro-3, Dro-5, and Dro-6 (Fig. 5b). However, although the same trend was observed for QTL Dro-1, Dro-4, and Dro-10, the differences in R 2 estimates were not statistically significant (Fig. 5b). On the other hand, QTL Dro-7, Dro-8, and Dro-9 showed little difference in R 2 estimates between the 57 and ≥124 populations, with the ≥124 populations resulting in slightly but not significantly larger R 2 estimates. Population size did not have significant effect on the statistical significance of LOD scores for QTL identified for maturity (Fig. 5a). However, there was significant inflation of the R 2 estimates based on smaller populations compared to those obtained using the 108 RIL population.
Discussion
Amplified fragment length polymorphism (AFLP) is a versatile technique that has been used successfully to study the genomic structure and construct genetic maps of many plant species using random or structured populations (Mohler and Schwarz 2004). Because no prior knowledge of genome sequence is required, AFLP technique facilitates studies in crops species that lag behind novel genome analysis techniques. Cowpea is among the “orphan” crops, which are crops of major economic importance in developing countries that lag behind other crops in genomic studies (Kaga et al. 2004; Nelson et al. 2004). As such, few genomic resources are available to facilitate genetic studies of important traits. The AFLP technique therefore, offers opportunity to generate quick and accurate genomic framework to facilitate such studies without major investment of time and resources. Previous studies have shown AFLP genotyping to be effective in cowpea. The most recent cowpea genetic linkage map is comprised of 267 AFLP, 133 RAPD, 36 RFLP, 3 morphological and 1 biochemical markers (Ouédraogo et al. 2002a). Other studies have utilized the AFLP technology to study genetic diversity (Tosti and Negri 2002; Gillaspie Jr et al. 2005; Fang et al. 2007) and develop molecular markers for resistance traits (Ouédraogo et al. 2002b; Li et al. 2007). In the current study, 378 polymorphic AFLP markers were generated of which 306 were successfully assigned to 11 linkage groups (Fig. 1). Linkage groups in this study were not aligned with previous maps and therefore their designations here have no correspondence to those described in previously published cowpea genetic maps. The AFLP map constructed in this study was significantly smaller (643 cM) than the MapMaker-generated 2,670-cM-long map reported by Ouédraogo et al. (2002a). Other previous cowpea genetic linkage maps of sizes 684 cM (Menancio-Hautea et al. 1993), 972 cM (Menéndez et al. 1997), and 669.8 cM (Ubi et al. 2000), were also significantly smaller than the Ouédraogo map. Although Gawłowska et al. (2005) showed that MapMaker and JoinMap software gave significantly different map sizes of the same mapping population with JoinMap giving the smaller version, differences between the map developed in the present study and the Ouédraogo map cannot be sufficiently explained by software differences. In addition, Menancio-Hautea et al. (1993), Menéndez et al. (1997), Ubi et al. (2000), and Ouédraogo et al. (2002a) used the MapMaker software but reported smaller map sizes. This suggests the influence of other factors such as differences in plant material and/or marker scoring. It is noteworthy that Ouédraogo et al. (2002a) identified a 580-cM-contiguous region composed entirely of AFLP markers on linkage group 1 of their map. This region had not been mapped in previous maps. The authors suggested that AFLP markers may have been more efficient in detecting polymorphisms in this genomic region compared to previous marker technologies. However, this finding has not been independently confirmed and given current genomic resources available in cowpea, these discrepancies cannot be adequately addressed. Future studies will need to facilitate alignment of these individual maps in order to resolve the disparity.
As observed in our earlier study (Muchero et al. 2008), genotypes IT93K503-1 and CB46 differed in their tolerance to seedling stage drought being tolerant and intolerant, respectively. A RIL population developed from the two genotypes segregated for these drought tolerance traits and evidence of transgressive segregants with extreme phenotypes for susceptibility and tolerance was found. Ten drought-related QTL were identified using this population. QTL mapping using the Kruskall–Wallis and MQM analysis yielded consistent results between experiments and across greenhouse and field environments. The stability of QTL expression across time and environments suggests that the genomic regions harboring the identified QTL carry genes that are of major importance in determining cowpea response to drought. Further, the fact that nine of the ten drought QTL and both maturity QTL were reliably detected with sufficient statistical significance using subsets of 51, 57, and 70 RILs suggests that the drought response and maturity mechanisms have high heritability. However, despite the statistical significance, the smaller population sizes resulted in a general inflation of R 2 estimates and lower LOD scores for both traits. This observation is in agreement with previous studies that have highlighted the increased risk for type II errors and inflation of percent phenotypic variance explained in small mapping populations (Vales et al. 2005). With the exception Dro-2 and Dro-6, the statistical significance of QTL described in this study was consistent across experiments. This provides evidence for the presence of genetic elements important for cowpea response to drought stress in these regions of the cowpea genome. However, the variability in R 2 estimates between different population sizes and also within experiments conducted with the same population size necessitate further studies in different genetic backgrounds to ascertain the magnitude of individual QTL contribution toward cowpea drought tolerance. In the absence of comparable studies in cowpea, the decision to calculate LOD significance threshold for individual linkage groups instead of adopting a genome-wide threshold may have been justified given the difference in thresholds obtained for larger compared to the smaller linkage groups. The highest achieved LOD threshold of 2.1 was considerably higher than the lowest 1.4 threshold obtained for linkage group 10. Therefore, adopting a genome-wide threshold may have resulted in the exclusion of QTL identified on smaller linkage groups 9 and 10 thereby increasing the risk of type II errors.
Phenotypes observed and assayed in response to both greenhouse and field drought stress support the senescence-based paradigm for drought response proposed previously (Muchero et al. 2008). As concluded in that study which characterized a diverse group of 14 cowpea genotypes, maintenance of stem greenness was highly correlated with drought tolerance. In the present study, visual scores of stem greenness identified the same QTL regions as recovery dry weight under greenhouse conditions. Visual scores of drought-induced senescence under field conditions were negatively correlated with greenhouse-based stem greenness and recovery dry weight and revealed the same suite of QTL as these greenhouse assays. This suggests that stem greenness and recovery dry weight are reliable greenhouse parameters that can be used to infer drought response under field conditions. In addition, visual scores of field drought-induced senescence revealed the same QTL as dry weight-based leaf damage assay used for evaluating drought-induced senescence under field conditions. These results provide confirmation that visual scores for drought-induced senescence are a reliable indicator of seedling drought tolerance that can be utilized in both greenhouse and field screening in cowpea. Similar results have been reported in sorghum (Sorghum bicolor L. Moench) with post-flowering drought tolerance mediated by the “stay-green” trait (Subudhi et al. 2000). Stay-green is a drought tolerance mechanism exhibited in some sorghum genotypes subjected to post-flowering drought stress. The trait allows tolerant genotypes to maintain green leaf area during the grain-filling stage, thereby allowing more productivity (Crasta et al. 1999). Numerous studies have mapped QTL involved in the stay-green trait on the sorghum genetic map (Crasta et al. 1999; Borrell et al. 2000; Subudhi et al. 2000; Tao et al. 2000; Sanchez et al. 2002). In most of these studies, visual scoring of premature leaf senescence was a reliable predictor of green leaf area at maturity (GLAM) in sorghum subjected to post-flowering drought stress (Crasta et al. 1999; Borrell et al. 2000; Subudhi et al. 2000; Tao et al. 2000). Using a sorghum recombinant inbred line population, Crasta et al. (1999) showed that genomic regions mediating the stay-green phenotype were distinct from regions mediating maturity, with the exception of one region where QTL for the two traits co-located. We have presented similar results for cowpea in this study, where we found drought-induced senescence QTL to be independent from maturity QTL.
Given current knowledge, it is not clear to what extent the sorghum stay-green trait, which is a post-flowering phenomenon, and the cowpea delayed drought-induced senescence trait, which we observed at the seedling stage, are related. The two may be the same phenomenon manifested in different plant growth stages. Regardless, the distinctness between maturity-related and drought-induced senescence has important implications for breeding strategies in cowpea. Early maturity is an important phenological trait in avoiding late-season drought stress because cultivars are able to complete their life cycle before the onset of severe drought conditions (Grantz and Hall 1982; Ehlers and Hall 1997). Our results suggested that it should be possible to pyramid early maturity with delayed drought-induced senescence to manage both early and late season drought stress in cowpea. Several of the recombinant inbred lines expressed the combined traits of early maturity with delayed drought-induced senescence.
Our study provides the first evidence of QTL mediating seedling drought response in cowpea and provides a platform for the identification of genetic factors determining this tolerance. A starting point may be pursuing the senescence-based model of seedling drought response in cowpea. Candidate pathways may be suggested from model plant systems where knowledge of stress-induced senescence is more advanced. Such candidate pathways may include the jasmonic acid and lipid signalling pathways that have been implicated in molecular responses of cowpea to drought stress. Previous studies using drought susceptible and drought tolerant cowpea genotypes demonstrated the differential expression of cDNA transcripts related to jasmonic acid biosynthesis and lipid hydrolysis under drought stress conditions. For example, phospholipase D (El-Maarouf et al. 1999) and galactolipid acyl hydrolase (Matos et al. 2001), which are both membrane lipid-degrading enzymes, were up-regulated in drought susceptible more than in drought tolerant cowpea genotypes. Further, the isolation of transcripts homologous to lipoxygenase (Iuchi et al. 1996a) and 12-oxophytodienoic acid reductase (Iuchi et al. 1996b) from cowpea leaves subjected to dehydration stress suggests a potential role for the jasmonic acid pathway in cowpea response to drought. This pathway reportedly mediates premature leaf senescence in Arabidopsis under abiotic stress (He et al. 2002). In addition, the ethylene pathway has been reported in numerous plant species to be involved in stress-induced leaf senescence (John et al. 1995; Young et al. 2004; Buchanan-Wollaston et al. 2005). Mapping genes associated with these pathways and investigating their co-localization with identified QTL may suggest candidate genes for further molecular studies.
Visual phenotyping for stem greenness under greenhouse conditions and for drought-induced senescence under field conditions should provide a rapid, reliable and cost effective alternative in screening and selecting for vegetative drought tolerance in applied cowpea breeding programs. Results of our study confirmed the applicability of this phenotyping process for identifying genomic regions associated with seedling drought tolerance. This suggests that selections based on these phenotypes should facilitate the successful introgression of genetic trait determinants of seedling drought tolerance into elite cowpea cultivars. Therefore, this study provides the basis for rapid screening of cowpea genotypes, as well as the development of molecular markers or identification of genes associated with seedling drought tolerance for use in marker-assisted selection, and breeding for drought tolerance in cowpea.
References
Analytical software (2003) Statistix 8 user’s manual. Analytical software, Tallahassee
Aswath CR, Kim SH, Mo SY, Kim DH (2005) Transgenic plants of creeping bent grass harboring the stress inducible gene, 9-cis-epoxycarotenoid dioxygenase, are highly tolerant to drought and NaCl stress. Plant Growth Regul 47:129–139
Borrell AK, Hammer GL, Douglas ACL (2000) Does maintaining green leaf area in sorghum improve yield under drought? I. Leaf growth and senescence. Crop Sci 40:1026–1037
Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin J-F, Wu S-H, Swidzinski J, Ishizaki K, Leaver CJ (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42:567–585
Contour-Ansel D, Torres-Franklin ML, Cruz de Carvalho MH, D’Arcy-Lameta A, Zuily-Fodil Y (2006) Glutathione reductase in leaves of cowpea: cloning of two cDNAs, expression and enzymatic activity under progressive drought stress, desiccation and abscisic acid treatment. Ann Bot 98:1279–1287
Crasta OR, Xu WW, Rosenow DT, Mullet J, Nguyen HT (1999) Mapping of post-flowering drought resistance traits in grain sorghum: association between QTLs influencing premature senescence and maturity. Mol Gen Genet 262:579–588
Curley J, Sim SC, Warnke S, Leong S, Barker R (2005) QTL mapping of resistance to gray leaf spot in ryegrass. Theor Appl Genet 111:1107–1117
D’Arcy-Lameta A, Ferrari-Iliou R, Contour-Ansel D, Pham-Thi A, Zuily-Fodil Y (2006) Isolation and characterization of four ascorbate peroxidase cDNAs responsive to water deficit in cowpea leaves. Ann Bot 97:133–140
Diop NN, Kidrič M, Repellin A, Gareil M, D’Arcy-Lameta A, Thi ATP, Zuily-Fodil Y (2004) A multicystanin is induced by drought-stress in cowpea (Vigna unguiculata (L.) Walp.) leaves. FEBS Lett 577:545–550
Doerge RW, Rebai A (1996) Significance thresholds for QTL interval mapping tests. Heredity 76:459–464
Ehlers JD, Hall AE (1997) Cowpea (Vigna unguiculata L. Walp.). Field Crops Res 53:187–204
El-Maarouf H, D’Arcy-Lameta A, Gareil M, Zuily-Fodil Y, Pham-Thi A (1999) Enzymatic activity and gene expression under water stress of phospholipase D in two cultivars of Vigna unguiculata L. Walp. differing in drought tolerance. Plant Mol Biol 39:1257–1265
El-Maarouf H, D’Arcy-Lameta A, Gareil M, Zuily-Fodil Y, Pham-Thi A (2001) Cloning and expression under drought of cDNAs coding for two PI-PLCs in cowpea leaves. Plant Physiol Biochem 39:167–172
Fang J, Devanand PS, Chao CT (2005) Practical strategy for identification of single nucleotide polymorphisms in fruiting Mei (Prunus mume Sieb. et Zucc.) from amplified fragment length polymorphism fragments. Plant Mol Biol Rep 23:227–239
Fang JC, Chao CT, Roberts PA, Ehlers JD (2007) Genetic diversity of cowpea [Vigna unguiculata (L.) Walp.] in four West African and USA breeding programs as determined by AFLP analysis. Genet Resour Crop Evol 54:1197–1209
Fery RL (2002) New opportunities in Vigna. In: Janick J, Whipkey A (eds) Trends in crops and new uses. ASHS press, Alexandria, pp 424–428
Gawłowska M, Święcicki W, Wolko B (2005) Comparison of genetic maps for two related pea populations (Pisum sativum L.). Pisum Genetics 37:19–23
Grantz DA, Hall AE (1982) Earliness of an indeterminate crop, Vigna unguiculata (L.) Walp., as affected by drought, temperature, and plant density. Aust J Agric Res 33:531–540
Gillaspie AG Jr, Hopkins MS, Dean RE (2005) Determining genetic diversity between lines of Vigna unguiculata subspecies by AFLP and SSR markers. Genet Resour Crop Evol 52:245–247
Hall AE (2004a) Comparative ecophysiology of cowpea, common bean, and peanut. In: Nguyen HT, Blum A (eds) Physiology and biotechnology integration for plant breeding. Marcel Dekker, Inc, New York, pp 271–326
Hall AE (2004b) Breeding for adaptation to drought and heat in cowpea. Eur J Agron 21:447–454
Hall AE, Cisse N, Thiaw S, Elawad HOA, Ehlers JD, Ismail AM, Fery R, Roberts PA, Kitch LW, Murdock LL, Boukar O, Phillips RD, McWatters KH (2003) Development of cowpea cultivars and germplasm by the Bean/Cowpea CRSP. Field Crops Res 82:103–134
He Y, Fukushige H, Hilderbrand DF, Gan S (2002) Evidence supporting the role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol 128:876–884
Iuchi S, Yamaguchi-Shinozaki K, Urao T, Shinozaki K (1996a) Characterization of two cDNAs for novel drought-inducible genes in the highly drought tolerant cowpea. J Plant Res 109:415–424
Iuchi S, Yamaguchi-Shinozaki K, Urao T, Terao T, Shinozaki K (1996b) Novel drought-inducible genes in the highly drought-tolerant cowpea: cloning of cDNAs and analysis of the expression of the corresponding genes. Plant Cell Physiol 37:1073–1082
Iuchi S, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K (2000) A stress-inducible gene for 9-cis-epoxycarotenoid dioxygenase involved in abscisic acid biosynthesis under water stress in drought-tolerant cowpea. Plant Physiol 123:553–562
John I, Drake R, Farrell A, Cooper W, Lee P, Horton P, Grierson D (1995) Delayed leaf senescence in ethylene-deficient ACC-oxidase antisense tomato plants: molecular and physiological analysis. Plant J 7(3):483–490
Kaga A, Vaughan DA, Tomooka N (2004) Molecular markers in Vigna improvement: understanding and using gene pools. In: Lörz H, Wenzel G (eds) Biotechnology in agriculture and forestry 55: molecular marker systems in plant breeding and crop improvement. Springer-Verlag, Berlin, pp 171–187
Kosambi DD (1944) The estimation of map distance from recombination values. Ann Eugen 12:172–175
Li G, Liu Y, Ehlers JD, Zhu Z, Wu X, Wang B, Lu Z (2007) Identification of an AFLP fragment linked to rust resistance in asparagus bean and its conversion to a SCAR marker. HortScience 42:1153–1156
Matkin OA, Chandler PA (1957) The U.C.-type soil mixes. In: Baker KF (ed) The U.C. system for producing healthy container-grown plants. Agricultural Publications, University of California Berkeley, pp 68–85
Matos AR, D’Arcy-Lameta A, França M, Pêtres S, Edelman L, Kader J, Zuily-Fodil Y, Pham-Thi A (2001) A novel patatin-like gene stimulated by drought stress encodes a galactolipid acyl hydrolase. FEBS Lett 491:188–192
Menancio-Hautea D, Fatokun CA, Kumar L, Danesh D, Young ND (1993) Comparative genome analysis of mungbean (Vigna radiata L. Wilczek) and cowpea (V. unguiculata L. Walpers) using RFLP mapping data. Theor Appl Genet 86:797–810
Menéndez CM, Hall AE, Gepts P (1997) A genetic linkage map of cowpea (Vigna unguiculata) developed from a cross between two inbred, domesticated lines. Theor Appl Genet 95:1210–1217
Mohler V, Schwarz G (2004) Genotyping tools in plant breeding: from restriction fragment length polymorphisms to single nucleotide polymorphisms. In: Lörz H, Wenzel G (eds) Biotechnology in agriculture and forestry 55: molecular marker systems in plant breeding and crop improvement. Springer-Verlag, Berlin, pp 26–38
Muchero W, Ehlers JD, Roberts PA (2008) Seedling stage drought-induced phenotypes and drought-responsive genes in diverse cowpea genotypes. Crop Sci 48:541–552
Nelson RJ, Naylor RL, Jahn MM (2004) The role of genomics research in improvement of “orphan” crops. Crop Sci 44:1901–1904
Ouédraogo JT, Gowda BS, Jean M, Close TJ, Ehlers JD, Hall AE, Gillaspie AG, Roberts PA, Ismail AM, Bruening G, Gepts P, Timko MP, Belzile FJ (2002a) An improved genetic linkage map for cowpea (Vigna unguiculata L.) combining AFLP, RFLP, RAPD, biochemical markers, and biological resistance traits. Genome 45:175–188
Ouédraogo JT, Tignegre J-B, Timko MP, Belzile FJ (2002b) AFLP markers linked to resistance against Striga gesnerioides race 1 in cowpea (Vigna unguiculata). Genome 45:787–793
Posthuma D, Luciano M, de Geus EJC, Wright MJ, Slagboom PE, Montgomery GW, Boomsma DI, Martin NG (2005) A genomewide scan for intelligence identifies quantitative trait loci on 2q and 6p. Am J Hum Genet 77:318–326
Sanchez AC, Subudhi PK, Rosenow DT, Nguyen HT (2002) Mapping QTLs associated with drought resistance in sorghum (Sorghum bicolor L. Moench). Plant Mol Biol 48:713–726
Sanginga N, Lyasse O, Singh BB (2000) Phosphorus use efficiency and nitrogen balance of cowpea breeding lines in a low P soil of the derived savanna zone in West Africa. Plant Soil 220:119–128
Subudhi PK, Rosenow DT, Nguyen HT (2000) Quantitative trait loci for the stay green trait in sorghum (Sorghum bicolor L. Moench): consistency across genetic backgrounds and environments. Theor Appl Genet 101:733–741
Tao YZ, Henzell RG, Jordan DR, Butler DG, Kelly AM, Mcintyre CL (2000) Identification of genomic regions associated with stay green in sorghum by testing RILs in multiple environments. Theor Appl Genet 100:1125–1232
Tosti N, Negri V (2002) Efficiency of three PCR-based markers in assessing genetic variation among cowpea (Vigna unguiculata subsp. unguiculata) landraces. Genome 45:268–275
Ubi EB, Mignouna H, Thottappilly G (2000) Construction of a genetic linkage map and QTL analysis using a recombinant inbred population derived from an intersubspecific cross of cowpea (Vigna unguiculata (L.) Walp.). Breed Sci 50:161–172
Vales MI, Schön CC, Capettini F, Chen XM, Corey AE, Mather DE, Mundt CC, Richardson KL, Sandoval-Islas JS, Utz HF, Hayes PM (2005) Effect of population size on the estimation of QTL: a test using resistance to barley stripe rust. Theor Appl Genet 111:1260–1270
Van Duivenbooden N, Abdoussalam S, Mohamed AB (2002) Impact of climate change on agricultural production in the Sahel-Part 2. Case study for groundnut and cowpea in Niger. Clim Change 54:349–368
Van Ooijen JW, Voorrips RE (2001) JoinMap 3.0: software for the calculation of genetic linkage maps. Plant Research International, Wageningen
Van Ooijen JW, Boer MP, Jansen RC, Maliepaard C (2002) MapQTL® 4.0 software for the calculation of QTL positions on genetic maps. Plant Research International, Wageningen
Visker MH, Keizer LC, Van Eck HJ, Jacobsen E, Colon LT (2003) Can the QTL for late blight resistance on potato chromosome 5 be attributed to foliage maturity type? Theor Appl Genet 106:317–325
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Vos P, Hogers R, Bleeker M, Reijans M, van de Le T, Hornes M, Frijiters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Watanabe S, Hakoyama S, Terao T, Singh BB (1997) Evaluation methods for drought tolerance of cowpea. In: Singh BB, Raj M, Dashiell KE, Jackai LEN (eds) Advances in cowpea research. Copublication of International Institute of Tropical Agriculture (IITA) and Japan International Research Center for Agricultural Sciences (JIRCAS). IITA, Ibadan, pp 87–97
Wittig R, König K, Schmidt M, Szarzynski J (2007) A study of climate change and anthropogenic impacts in West Africa. Environ Sci Pollut Res 14:182–189
Young TE, Meeley RB, Gallie DR (2004) ACC synthase expression regulates leaf performance and drought tolerance in maize. Plant J 40:813–825
Acknowledgements
The work was funded in part by Bean/Cowpea Collaborative Research Support Program, USAID Grant no. GDG-G-00-02-00012-00 and the CGIAR Generation Challenge Program. The opinions and recommendations herein are those of the authors and not necessarily those of USAID.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J.-L. Jannink.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Muchero, W., Ehlers, J.D., Close, T.J. et al. Mapping QTL for drought stress-induced premature senescence and maturity in cowpea [Vigna unguiculata (L.) Walp.]. Theor Appl Genet 118, 849–863 (2009). https://doi.org/10.1007/s00122-008-0944-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-008-0944-7