Abstract
Hypersensitive resistance (HR) is an efficient defense strategy in plants that restricts pathogen growth and can be activated during host as well as non-host interactions. HR involves programmed cell death and manifests itself in tissue collapse at the site of pathogen attack. A novel hypersensitivity gene, Ny-1, for resistance to Potato virus Y (PVY) was revealed in potato cultivar Rywal. This is the first gene that confers HR in potato plants both to common and necrotic strains of PVY. The locus Ny-1 mapped on the short arm of potato chromosome IX, where various resistance genes are clustered in Solanaceous genomes. Expression of HR was temperature-dependent in cv. Rywal. Strains PVYO and PVYN, including subgroups PVYNW and PVYNTN, were effectively localized when plants were grown at 20°C. At 28°C, plants were systemically infected but no symptoms were observed. In field trials, PVY was restricted to the inoculated leaves and PVY-free tubers were produced. Therefore, the gene Ny-1 can be useful for potato breeding as an alternative donor of PVY resistance, because it is efficacious in practice-like resistance conferred by Ry genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants have evolved complex genetic systems to defend against pathogen attack. Elucidation of the molecular mechanism of plant disease resistance is important for several reasons. First, understanding how plants respond to pathogens is critical for the application of engineering for increased crop resistance to viruses, viroids, bacteria and fungi. In addition, the determination of the pathway from initial recognition of a pathogen to the induction defence-strategy will yield insight into basic aspects of specific signal transduction pathways.
Potato virus Y (PVY) is a member of the genus Potyvirus (Potyviridae family). Two main groups of PVY strains, PVY0 and PVYN, separated according to their pathogenic properties, are the most harmful viruses in cultivated potato (Solanum tuberosum ssp. tuberosum). PVY0 (common, ordinary strain) induces leaf mottling, extensive necrotic and drop streak symptoms, and often leads to plant death. PVYN (tobacco veinal necrosis strain) causes, in general, mild mosaic symptoms in potato leaves (De Bokx and Huttinga 1981; Brunt 2001; Valkonen 2007).
A subgroup of PVYN, designated as PVYNTN, is responsible for potato tuber necrotic ringspot disease, which results in losses of tuber quality (Beczner et al. 1984). The second subgroup, PVYNW, originally described in Polish cv. Wilga, as PVYNWi (Chrzanowska 1991), induces vein necrosis in Nicotiana tabaccum cv. Samsun and mild mosaic in potato plants, but is serologically related to PVY0 (Chrzanowska 1994), probably as the outcome of recombination events in the virus coat protein gene (Glais et al. 2005).
In potato, there are two main types of resistance to PVY, extreme resistance (ER) and hypersensitive resistance (HR). The Ry genes for ER confer extremely high level of protection against different strains of PVY (Ross 1986; Valkonen et al. 1996). Three Ry genes have been localized by molecular mapping on potato chromosomes. Ry sto (designated also as Ry-f sto ) derived from S. stoloniferum and Ry adg from S. tuberosum ssp. andigena mapped on potato chromosomes XII (Flis et al. 2005; Song et al. 2005) and XI (Hämäläinen et al. 1997), respectively. The gene Ry chc derived from S. chacoense mapped to the distal end of potato chromosome IX (Sato et al. 2006). The gene Ry sto was also localized on chromosome XI (Brigneti et al. 1997), in the same region, where Ry adg was mapped (Gebhardt and Valkonen 2001). Recently, however, the Ry adg -linked markers used in the S. stoloniferum mapping experiments did not confirm the position of Ry sto on potato chromosome XI (Valkonen et al. 2007).
The HR to PVY is strain specific in potato. Hypersensitivity to PVY0 and/or PVYN was described in wild Solanum species (Valkonen 1997; Ruiz de Galarreta et al. 1998; Solomon-Blackburn and Barker 2001). HR was also observed in cultivated potato, however, only after infection with the ordinary strain of PVY (Jones 1990; Valkonen et al. 1998; Sorri et al. 1999). Potato cultivars expressing HR to PVYN infection were not reported so far (Valkonen 2007). The first HR gene, Ny tbr , causing necrotic response to PVY0 infection in potato mapped on potato chromosome IV (Celebi-Toprak et al. 2002). In this paper, we report the first potato HR gene, which induces necrotic response and restriction of common and necrotic variants of PVY. The gene, designated as Ny-1, mapped on potato chromosome IX.
Materials and methods
Plant material and PVY strains
The tetraploid potato (Solanum tuberosum ssp. tuberosum, 2n = 4x = 48) population ‘RxA’, consisting of 200 F1 individuals, was obtained from a cross between Rywal, a cultivar with HR to PVY and the susceptible cv. Accent. Rywal originated from a cross of Polish cv. Dalia and Polish clone DM 1696. This clone was developed from Dutch cv. Edzina × Polish clone DM 51/5-7 cross. Further pedigree information for cultivars Dalia and Edzina, and clone DM 51/5-7 are compiled in Świeżyński et al. (1997) and the potato pedigree database (http://www.dpw.wau.nl/pv). Four PVY variants: PVY0LW, PVYNNy, PVYNW and PVYNTNBo, which derived from the corresponding potato cvs: Lipiński Wczesny, Nysa, Wilga and Bona, were multiplied and maintained in cv. Samsun tobacco plants.
Resistance assays for PVY strains
Two types of assays were performed to evaluate the resistance of cvs Rywal and Accent to PVY strains. Plants were grown for 2 weeks in the greenhouse and transferred 1 week before the infection experiments to growth chambers with controlled environmental conditions (20 or 28°C, 16 h light, 100 μm/s/m2, 8 h dark). For each cultivar, six detached leaves or six plants were mechanically inoculated with a sap extracted from the tobacco plants, infected with PVY0LW, PVYNNy, PVYNW or PVYNTNBo. Inoculated detached leaves and plants were divided into two groups. One group was incubated at 20°C, the other at 28°C. Water-treated detached leaves and plants were used as controls. Inoculated detached leaves were placed on a wet filter paper in two plastic trays, which were covered with glass plates to maintain high humidity. Hypersensitivity was visualized after 4–6 days. Nine days post-inoculation, 1 g samples of inoculated and non-inoculated upper leaves from each of the plants were collected in order to detect the virus by RT-PCR.
For the mapping experiments, two tuber-derived plants per clone of each F1 individuals of the ‘RxA’ population were screened for HR to PVYNW and PVYNTNBo infection at 20°C. In addition, three corresponding detached leaves were scored by inoculation with PVYNTNBo. Plants which developed necrotic symptoms were classified as resistant.
Detection of PVY RNA by RT-PCR
RNA was isolated using TRIZOL method (Chomczyński and Sacchi 1987). Reverse-transcription reactions contained 1 μg of DNase I treated RNA from PVY or mock inoculated plants. The reactions were primed with oligo(dT) and elongated by the PVY specific sequence (5′oligo(dTx30)GTCTCCTGATTGAAGTTTACAG3′). 200 U M-MuLV Reverse Transcriptase (Fermentas, Vilnius, Lithuania) were used to synthesize first-strand cDNA. PCR amplification from PVY coat protein RNA (PVYCP) was performed with the PVYCPf and PVYCPr primers for 30 cycles at 94°C for 30 s, 53°C for 30 s, and 72°C for 45 s. The primer sequences were: PVYCPf, 5′AAATACTCGGGCAACTCAATCACA3′; PVYCPr, 5′CACCGTCCAACCCGAAAAG3′. They were based on coat protein sequences for various PVY strains, available in GenBank database. PrimerSelect software from Lasergene 6.1 set (DNASTAR Inc., Madison WI, USA) was used for primer design.
Plant DNA isolation, PCR, electrophoresis, DNA cloning and sequencing
Extraction of plant genomic DNA and electrophoresis were performed as previously described (Flis et al. 2005). Inter-simple sequence repeat (ISSR) markers were amplified in 20 μl of 20 mM Tris–HCl pH 8.4, 50 mM KCl, 2% DMSO, 2.5 mM MgCl2, 0.2 mM of each deoxynucleotide, 0.3 μM of primer, containing 1 U Taq DNA Polymerase (Invitrogen, Carlsbad, CA, USA) and 30 ng genomic DNA as template. The PCR parameters were: 40 cycles of 94°C for 20 s, 42°C for 30 s, 72°C for 2 min and a final extension time of 10 min at 72°C. The ISSR marker UBC8951200 was cloned and sequenced as previously described (Marczewski et al. 2006) and compared to the NCBI sequence database with the BLASTN program.
Three markers: sequence characterized amplified region (SCAR) SC8951139 (NCBI GenBank accession EF555209), GP41443 (AJ487356) and conserved ortholog set II (COSII) C2_At3g168401100 (SGN-U284031) were amplified in 20 μl of 20 mM Tris–HCl pH 8.4, 50 mM KCl, 1.5 mM MgCl2, 0.1 mM of each deoxynucleotide, 0.2 μM of each primer, containing 1 U Taq DNA Polymerase (Invitrogen) and 30 ng genomic DNA as template. Forward and reverse primers, designed as described in Marczewski et al. (2006), were: f: 5′GGTAGCTCTTGATCTCGTCTT3′, r: 5′GTAGCTCTTGATCACCCATTT3′ for SC895; f: 5′GTTGGTACCAGGCTTGTT3′, r: 5′CATTCGGTGCTTTAGGAT3′ for GP41 and f: 5′TCCAGTGTCCAAAGAAAGAAAA3′, r: 5′ATGCTCATGTCCCCGAAACC3′ for C2_At3g16840. The PCR parameters for amplifying SC8951139 and C2_At3g168401100 were: 94°C for 60 s followed by 40 cycles of 93°C for 25 s, 56°C for 35 s, 72°C for 90 s and a final extension time of 5 min at 72°C. GP41443 was amplified at the annealing temperature of 54°C.
Detection of PVY in field-grown plants
Four groups of cv. Rywal plants, 60 plants each, were subjected to PVY-exposure field trials at Młochów, during five vegetation seasons, 1999–2003. One group consisted of plants grown from the virus-free tubers, whereas in the other groups three successive tuber progenies were planted. Each third row of the field trials was planted with tubers of PVY-infected potato plants. In addition, during each year 120 plants grown from field tubers harvested in the northeastern region of Poland were planted in a greenhouse at Młochów. The presence of PVY was tested in leaves of 5- to 6-week-old plants by enzyme-linked immunosorbent assay (ELISA) using the monoclonal cocktail Bioreba AG kit (Reinach, Switzerland).
Mapping of the locus Ny-1
The diploid mapping population ‘Erwinia’ (Zimnoch-Guzowska et al. 2000) was used to map the SCAR marker SC8951139 linked to the gene Ny-1. SC8951139, amplified from parental clone DG 81-68, was scored as presence or absence of the marker fragment in 78 F1 progeny. The map position was identified relative to the amplified fragment length polymorphism (AFLP) and restriction fragment length polymorphism (RFLP) map existing for this population using the software package MAPRF (E. Ritter, NEIKER, 01080 Victoria, Spain). The position of Ny-1 in cv. Rywal was confirmed in the population ‘RxA’ by detecting linkage to the anchor markers GP41 and C2_At3g16840, specific for potato chromosome IX (PoMaMo database, http://gabi.rzpd.de/database/maps.shtml) and tomato chromosome 9 (Tomato-EXPEN 2000 map, http://www.sgn.cornell.edu/search/markers/markersearch.pl), respectively.
Results
Reaction of cvs Rywal and Accent to infection with PVY strains
When detached leaves or whole plants of PVY susceptible cv. Accent were mechanically inoculated with strains PVYOLW, PVYNNy, PVYNW and PVYNTNBo, no symptoms were observed at 20 or 28°C (not shown). The systemic, symptomless infection was also associated with plants of cv. Rywal inoculated and grown at 28°C. In contrast, at 20°C, necrotic lesions were observed in leaves of cv. Rywal, 4–6 days post-inoculation for each strain (Fig. 1). The viral RNA was not detected in the upper, non-inoculated leaves of cv. Rywal collected 9 days after inoculation (Table 1). Increasing the temperature from 20 to 28°C allowed the virus to occasionally move from inoculated leaves to leaves on upper parts of the plant, which remained symptom-free but contained detectable amounts of PVY. The systemic virus spread did not occur when the inoculated leaves were detached from the plant prior to their transfer to the higher temperature (not shown). However, during 5 years of experiments for the field-grown plants, no more than occasional systemic infections with PVY, resulting in plant death, were observed.
Mapping of the gene Ny-1
Hypersensitive resistance was observed in 93 F1 hybrids of the mapping population ‘RxA’ using both the detached leaf and the whole plant-inoculation tests. The PVY susceptible group consisted of 107 F1 hybrids. The segregation ratio of 1:1 (χ 2 = 0.98, P = 0.32) fits the genetic model of a single, dominant gene for HR to PVY in simplex state in cv. Rywal.
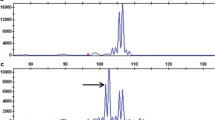
Seventy-eight ISSR primers, that were informative in previous studies (Flis et al. 2005), were tested by PCR in the parental DNA samples. Of 16 PCR products generated specifically in the resistant parent cv. Rywal, only one, a 1,200 bp long DNA fragment amplified using primer UBC895 (5′AGAGTTGGTAGCTCTTGATC3′) was seen as a faint band (Fig. 2, lane 1) in a bulked DNA sample constructed from eight HR progeny. The marker was not observed in the susceptible DNA bulk. UBC8951200 was cloned and sequenced in order to obtain more reliable and specific PCR amplification. Based on the cloned 1,152 bp marker, which revealed no apparent homology to any sequences available in GenBank, the SCAR marker SC8951139 (EF555209) was developed as the clear, strong PCR product, amplified in cv. Rywal (Fig. 2, lane 3). SC8951139 was used to screen the 200 F1 individuals of the ‘RxA’ family. The marker fragment was observed in all 93 resistant and in 1 susceptible F1 plant, indicating a close linkage (0.5 cM) between the SCAR marker and the locus Ny-1. Segregation of SC8951139 in the ‘Erwinia’ mapping population (Zimnoch-Guzowska et al. 2000) revealed the map position of Ny-1 on the short arm of potato chromosome IX. The chromosome location was confirmed when a 443 bp sequence tagged site (STS) amplicon (Fig. 2, lane 5) of the chromosome IX specific marker GP41 (http://gabi.rzpd.de/database/maps.shtml) was positioned 1 cM from Ny-1 in cv. Rywal. The locus Ny-1 was also found to be separated by 6 cM from the COSII marker C2_At3g16840 described in the reference Tomato Genetic Map at position 88 cM on tomato chromosome 9 (Wu et al. 2006). C2_At3g16840 was scored based on two restriction fragments of 350 and 500 bp, generated after TaqI digestion of the approximately 1,100 bp amplicon (Fig. 2, lane 7). The marker GP41 was found in the Tomato-EXPEN 1992 genetic map at position 104.8 cM on chromosome 9 (http://www.sgn.cornell.edu/cview). The syntenic relationships between tomato and potato genomes implicate, therefore, the proximal orientation of C2_At3g16840 and the distal locations of SC8951139 and GP41 in the linkage group IX (Fig. 3).
Patterns of amplified DNA of resistant parent cv. Rywal (lanes 1, 3, 5 and 7) and susceptible parent Accent (lanes 2, 4, 6 and 8), indicating the ISSR marker UBC8951200 (lane 1), the STS marker SC8951139 (lane 3), the SCAR marker GP41443 (lane 5) and the COSII marker C2_At3g16840 after digestion with TaqI (lanes 7 and 8). Arrows point to the marker products that were scored. Lanes M contain the 250-bp (upper) and 100-bp (lower) DNA ladders as molecular size markers
Discussion
Plant resistance is often accompanied by rapid tissue necrotization at pathogen entry sites, referred to as HR. High temperature may induce a switch from necrotic reactions to absence or mild systemic symptoms in virus infected plants (Malamy et al. 1992; Zheng et al. 2005). For example, HR to PVYN in S. sparsipilum and S. sucrense and to PVY0 in potato cv. Pito was efficiently expressed at temperatures 16–18°C, indicated by the local and/or systemic necrosis, while at higher temperatures (19/24°C), only leaf drop and mosaic symptoms were developed (Valkonen 1997). The temperature-sensitive response was also observed in potato plants with the genes Nx and Nb, which induced HR to Potato virus X (PVX) infection (Adams et al. 1986). HR induced by Ny-1 in cv. Rywal was efficiently developed at 20°C, whereas local necrotic lesions were not revealed in plants grown constitutively at elevated (28°C) temperature. Moreover, this temperature allowed the plants to be systemically infected with PVY0LW, PVYNNy, PVYNW or PVYNTNBo with the absence of viral symptom expression. In Poland, potato cultivars are grown between April and August, when daily minimum and maximum temperatures may reach the values below 10°C and above 35°C. However, throughout 5 years of the field experiments at Młochów plants of cv. Rywal produced PVY-free tubers. Młochów is located in the central region of Poland, where PVY infection pressure is intense (Gabriel 1995). PVY was also not revealed in tubers collected in the northeastern region of Poland. Moreover, during several years of cv. Rywal reproduction at the Plant Breeding Station Szyldak, northern Poland, harvested tubers were PVY-free (R. Bruski, personal communication). It is possible that the natural PVY infection pressure caused by aphids, which transmit PVY non-persistently, was too slow to break the virus restriction at the site of infection and activate its systemic movement in Ny-1 possessing plants, even at the higher temperatures. To our knowledge cv. Rywal is the first commercial potato cultivar, in which HR to both common and necrotic PVY strains, including PVYNW and PVYNTN, was effectively expressed.
The extreme resistance gene Ry chc mapped recently on potato chromosome IX in Japanese cv. Konafubuki was tightly linked to the marker TG421 (Sato et al. 2006). The Ny-1-linked marker C2_At3g16840 was found to be linked to TG421, within 5 cM, in the reference Tomato-EXPEN 2000 map (http://www.sgn.cornell.edu/search/markers/markersearch.pl). Therefore, Ry chc and Ny-1 may be new members of the resistance hot spot on potato chromosome IX, in which resistance genes and quantitative trait loci to various pathogens have been previously described (Marczewski et al. 2006; Rouppe van der Voort et al. 2000; Simko et al. 2004; Smilde et al. 2005; Śliwka et al. 2006; Tommiska et al. 1998; Zimnoch-Guzowska et al. 2000).
Ry genes for extreme resistance to PVY Ry sto have been introgressed mainly into European potato cultivars (Flis et al. 2005; Song et al. 2005). However, ER cultivars and breeding lines often exhibit male sterility (Lössl et al. 2000), which limits their usefulness in potato breeding programs. Cultivars and breeding clones derived from Polish, German, Dutch and North American gene pools are in the pedigree of cv. Rywal. Hypothetically, some of them can possess the gene Ny-1, and the Ny-1-linked markers should be helpful for their selection. Therefore, we postulate that Ny-1, which provides a level of the field resistance to PVY as satisfactory as the Ry genes, may be available and useful in several potato breeding programs.
References
Adams SE, Jones RAC, Coutts RHA (1986) Effect of temperature on potato virus X infection in potato cultivars carrying different combinations of hypersensitivity genes. Plant Pathol 35:517–526
Beczner L, Horvath J, Romhanyi I, Forster H (1984) Studies on the etiology of tuber necrotic ringspot disease in potato. Potato Res 27:339–352
Brigneti G, Garcia-Mas J, Baulcombe DC (1997) Molecular maping of the potato virus Y resistance gene Ry sto in potato. Theor Appl Genet 94:198–203
Brunt AA (2001) Potyviruses. In: Loebenstein G, Berger PH, Brunt AA, Lawson RH (eds) Virus and virus-like diseases of potatoes and production of seed-potatoes. Kluwer, Dordrecht, Boston, London, pp 77–84
Celebi –Toprak F, Slack SA, Jahn MM (2002) A new gene, Ny tbr , for hypersensitivity to Potato virus Y from Solanum tuberosum maps to chromosome IV. Theor Appl Genet 104:669–674
Chomczyński P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159
Chrzanowska M (1991) New isolates of the necrotic strain of potato virus Y (PVNN) found recently in Poland. Potato Res 34:179–182
Chrzanowska M (1994) Differentiation of potato virus Y (PVY) isolates. Phytopathol Pol 8:15–20
De Bokx JA, Huttinga H (1981) Potato virus Y. CMI/AAB Description of plant viruses no. 242
Flis B, Hennig J, Strzelczyk-Żyta D, Gebhardt C, Marczewski W (2005) The Ry-f sto gene from Solanum stoloniferum for extreme resistant to Potato virus Y maps to potato chromosome XII and is diagnosed by PCR marker GP122718 in PVY resistant potato cultivars. Mol Breed 15:95–101
Gabriel W (1995) Une méthode d’utilisation possible de l’échelle de 1 á 9 pour l’évaluation de la résistance aux virus chez les cultivars de pomme de terre. Potato Res 38:297–305
Gebhardt C, Valkonen JPT (2001) Organization of genes controlling disease resistance in the potato genome. Annu Rev Phytopathol 39:79–102
Glais L, Tribodet M, Kerlan C (2005) Specific detection of the PVYN-W variant of Potato virus Y. J Virol Methods 125:131–136
Hämäläinen JH., Watanabe KN, Valkonen JPT, Arihara A, Plaisted RL, Pehu E, Miller L, Slack SA (1997) Mapping and marker-assisted selection for a gene for extreme resistance to potato virus Y. Theor Appl Genet 94:192–197
Jones RAC (1990) Strain group specific and virus specific hypersensitive reactions to infection with potyviruses in potato cultivars. Ann Appl Biol 117:93–105
Lössl A, Götz M, Braun A, Wenzel G (2000) Molecular markers for cytoplasm in potato: male sterility and contribution of different plastid-mitochondrial configurations to starch production. Euphytica 116:221–230
Malamy J, Hennig J, Klessig DF (1992) Temperature-dependent induction of salicylic acid and its conjugates during the resistance response to tobacco mosaic virus infection. Plant Cell 4:359–366
Marczewski W, Strzelczyk-Żyta D, Hennig J, Witek K, Gebhardt C (2006) Potato chromosomes IX and XI carry genes for resistance to potato virus M. Theor Appl Genet 112:1232–1238
Rouppe van der Voort J, van der Vossen E, Bakker E, Overmars H, van Zandvoort P, Hutten R, Klein-Lankhorst R, Bakker J (2000) Two additive QTLs conferring broad-spectrum resistance in potato to Globodera pallida are localized on resistance gene clusters. Theor Appl Genet 101:1122–1130
Ross H (1986) Potato breeding—problems and perspectives. J Plant Breed Suppl 13
Ruiz de Galarreta JI, Carrasco A, Salazar A, Barrena I, Iturritxa E, Marqinez R, Legorburu F J, Ritter E (1998) Wild Solanum species as resistance sources against different pathogens of potato. Potato Res 41:57–68
Sato M, Nishikawa K, Komura K, Kosaka K (2006) Potato virus Y resistance gene, Ry chc , mapped to the distal end of potato chromosome 9. Euphytica 149:367–372
Simko I, Constanzo S, Haynes KG, Christ BJ, Jones RW (2004) Linkage disequilibrium mapping of a Verticilium dahliae resistance quantitative trait locus in tetraploid potato (Solanum tuberosum) through a candidate gene approach. Theor Appl Genet 108:217–224
Smilde WD, Brigneti G, Jagger L, Perkins S, Jones JDG (2005) Solanum mochiquense chromosome IX carries a novel late blight resistance gene Rpi-moc1. Theor Appl Genet 110:252–258
Solomon-Blackburn RM, Barker H (2001) A review of host major-gene resistance to potato viruses X, Y, A and V in potato: genes, genomics and mapped locations. Heredity 86:8–16
Song YS, Hepting L, Schweizer G, Hartl L, Wenzel G, Schwarzfischer A (2005) Mapping of extreme resistance to PVY (Ry sto ) on chromosome XII using anther-culture-derived primary dihaploid potato lines. Theor Appl Genet 111:879–887
Sorri VA, Watanabe KN, Valkonen JPT (1999) Predicted kinase-3a motif of a resistance gene analogue as a unique marker for virus resistance. Theor Appl Genet 99:164–170
Śliwka J, Jakuczun H, Lebecka R, Marczewski W, Gebhardt, Zimnoch-Guzowska E (2006) A novel late blight resistance gene Rpi-phu1 mapped to potato chromosome IX is not related to long vegetation period. Theor Appl Genet 113:685–695
Świeżyński KM, Haynes KG, Hutten RCB, Sieczka MT, Watts P, Zimnoch-Guzowska E (1997) Pedigree of European and North-American potato varieties. Plant Breed Seed Sci 41(1 supplement):3–149
Tommiska TJ, Hämäläinen JH, Watanabe KN, Valkonen JPT (1998) Mapping of the gene Nxphu that controls hypersensitive resistance to potato virus X in Solanum phureja IvP35. Theor Appl Genet 96:840–843
Valkonen JPT (1997) Novel resistances to four potyviruses in tuber-bearing potato species, and temperature-sensitive expression of hypersensitive resistance to potato virus Y. Ann Appl Biol 130:91–104
Valkonen JPT (2007) Viruses: economical losses and biotechnological potential. In: Vreugdenhil D, Bradshaw J, Gebhardt C, Govers F, Taylor M, MacKerron D, Ross H (eds) Potato biology and biotechnology: advances and perspectives, Elsevier, Amsterdam, pp 619–641
Valkonen JPT, Jones RAC, Slack SA, Watanabe KN (1996) Resistance specificity to viruses in potato: standardization of nomenclature. Plant Breed 115:433–438
Valkonen JPT, Rokka V-M, Watanabe KN (1998) Examination of the leaf-drop symptom of virus-infected potato using anther culture-derived haploids. Phytopathology 88:1073–1077
Valkonen JPT, Wiegmann K, Hämäläinen JH, Marczewski W, Watanabe KN (2007) Evidence for utility of the same PCR-based markers for selection of extreme resistance to Potato virus Y controlled by Ry sto of Solanum stoloniferum derived from different sources. Ann Appl Biol (in press)
Wu F, Mueller LA, Crouzillat D, Pétiard V, Tanksley SD (2006) Combining bioinformatics and phylogenetics to identify large sets of single-copy orthologous genes (COSII) for comparative, evolutionary and systematic studies: a test case in the euasterid plant blade. Genetics 174:1407–1420
Zheng C, Chen P, Gergerich R (2005) Effect of temperature on the expression of necrosis in soybean infected with Soybean mosaic virus. Crop Sci 45:916–922
Zimnoch-Guzowska E, Marczewski W, Lebecka R, Flis B, Schafer-Pregl R, Salamini F, Gebhardt C (2000) QTL analysis of new sources of resistance to Erwinia carotovora ssp. atroseptica in potato done by AFLP, RFLP, and resistance-gene-like markers. Crop Sci 40:1156–1167
Acknowledgments
We thank Bogdan Flis and Ewa Zimnoch-Guzowska for helpful comments on the manuscript and Andrzej Pałucha for the oligo(dT)-PVY primer sequence. The research was supported by the State Committee for Scientific Research in Poland, project PBZ/KBN/110/PO4/2004.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. Waugh.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Szajko, K., Chrzanowska, M., Witek, K. et al. The novel gene Ny-1 on potato chromosome IX confers hypersensitive resistance to Potato virus Y and is an alternative to Ry genes in potato breeding for PVY resistance. Theor Appl Genet 116, 297–303 (2008). https://doi.org/10.1007/s00122-007-0667-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-007-0667-1