Abstract
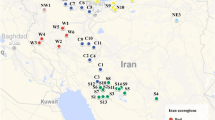
Characterizing and inferring the buffalograss [Buchloe dactyloides (Nutt.) Engelm.] genome organization and its relationship to geographic distribution are among the purposes of the buffalograss breeding and genetics program. This buffalograss study was initiated to: (1) better understand the buffalograss ploidy complex using various marker systems representing nuclear and organelle genomes; (2) determine whether the geographic distribution was related to nuclear and organelle genome variation; and (3) compare the genetic structure of accessions with different ploidy levels. The 20 buffalograss genotypes (15 individuals from each genotype) that were studied included diploid, tetraploid, pentaploid, and hexaploid using nuclear (intersimple sequence repeat (ISSRs), simple sequence repeat (SSRs), sequence related amplified polymorphism (SRAPs), and random amplified polymorphic DNA (RAPDs)) and cytoplasmic markers (mtDNA and cpDNA). There was a significant correlation between the ploidy levels and number of alleles detected using nuclear DNA (ISSR, SSR, and SRAP, r=0.39, 0.39, and 0.41, P<0.05, respectively), but no significant correlation was detected when mitochondrial (r=0.17, P<0.05) and chloroplast (r=0.11, P<0.05) DNA data sets were used. The geographic distribution of buffalograss was not correlated with nuclear and organelle genome variation for the genotypes studied. Among the total populations sampled, regression analysis indicated that geographic distance could not explain genetic differences between accessions. However, genetic distances of those populations from the southern portion of buffalograss adaptation were significantly correlated with geographic distance (r= 0.48, P<0.05). This result supports the hypothesis that genetic relationship among buffalograss populations cannot be estimated based only on geographic proximity.
Similar content being viewed by others
References
Allouis S, Qi X, Lindup S, Gale MD, Devos K M (2001) Construction of BAC library of pearl millet, Pennisetum glaucum. Theor Appl Genet 102:1200–1205
Arrieta-Montiel M, Lyznik A, Woloszynska M, Janska H, Tohme J, Mackenzie S (2001) Tracing evolutionary and developmental implications of mitochondrial stoichiometric shifting in the common bean. Genetics 158:851–864
Beard JB (1973) Turfgrass: science and culture. Prentice-Hall, Englewood Cliffs, NJ
Budak H, Pedraza F, Baenziger PS, Cregan PB, Dweikat I (2003) Development and utilization of SSR to estimate genetic diversity in a collection of pearl millet germplasm. Crop Sci 43:2284–2290
Budak H, Shearman RC, Parmaksiz I, Gaussoin RE, Riordan TP, Dweikat I (2004a) Molecular characterization of buffalograss germplasm using sequence related amplified polymorphism markers. Theor Appl Genet 108: 328–334
Budak H, Shearman RC, Parmaksiz I, Dweikat I (2004b) Comparative analysis of seeded and vegetative buffalograsses based on phylogenetic relationship using ISSR, SSR, RAPD and SRAP. Theor Appl Genet 109: 280–288
Budak H, Shearman RC, Dweikat I (2004c) Cloning and characterization of resistance gene like sequences in warm season turfgrass species. In: Proceedings of the international conference on mathematics and engineering techniques in medicine and biological science. Las Vegas, pp 225–230
Budak H, Shearman RC, Dweikat I (2005) Evolution of Buchloë dactyloides based on cloning and sequencing of matK, rbcL, and cob genes from plastid and mitochondrial genomes. Genome 48:411–416
Corderio GM, Taylor GO, Henry RJ (2000) Characterization of microsatellite markers from sugarcane (Saccharum sp.), a highly polyploidy species. Plant Sci 155:161–168
Excoffier L, Smouse P, Quattro J (1992) Analysis of molecular variance inferred for metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Grant V (1981) Plant speciation, 2nd edn. Columbia University Press, New York
Grivet G, Heinze B, Vendramin GG, Petit RJ (2001) Genome walking with consensus primers: application to the large single copy region of chloroplast DNA. Mol Ecol Notes 1:345–349
Hitchcock AS (1951) Manual of the grasses of the United States, 2nd edn. US Department of Agriculture Misc Pub 200., Washington
Huff DR, Peakall R, Smouse PE (1993) RAPD variation within and among natural populations of outcrossing buffalograss [Buchloë dactyloides (Nutt) Engelm]. Theor Appl Genet 86:927–934
Huff DR, Quinn JA, Higgins B, Palazzo AJ (1998) Random amplified polymorphic DNA (RAPD) variation among native little bluestem [Schizachyrium scoparium (Michx.) Nash] populations from sites of high and low fertility in forest and grassland biomes. Mol Ecol 7:1591–1597
Johnson PG, Riordan TP, Arumuganathan K (1998) Ploidy level determinations in buffalograss clones and populations. Crop Sci 38:478–482
Johnson PG, Kenworthy KE, Auld DL, Riordan TP (2001) Distribution of buffalograss polyploid variation in the southern Great Plains. Crop Sci 41:909–913
Kellogg EA, Appels R, Mason-Gramer RJ (1996) When gene trees tell different stories: the diploid genera of Triticeae (Gramineae). Syst Bot 21:321–347
Leitch I, Chase MW, Bennett MD (1998) Phylogenetic analysis of DNA C-values provides evidence for a small ancestral genome size in flowering plants. Ann Bot 82:85–94
Li G, Quiros CF (2001) Sequence-related amplified polymorphism (SRAP), a new marker system based on a sample PCR reaction: its application to mapping and gene tagging in Brassica. Theor Appl Genet 103: 455–461
Mason-Gamer RJ, Kellogg EA (1996a) Chloroplast DNA analysis of the monogenomic Triticeae: phylogenetic implications and genome specific markers. In: Jauhar PP (ed) Methods of genome analysis in plants. CRC Press, Boca Raton, pp 301–325
Mason-Gamer RJ, Kellogg EA (1996b) Testing for phylogenetic conflict among molecular data sets in tribe Triticeae (Gramineae). Syst Biol 45:524–545
Masterson J (1994) Stomatal size in fossil plants:evidence for polploidy in majority of angiosperms. Science 264:421–424
Nei M, and Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Ortmann J, Schacht WH, Stubbendieck J, Brink D (1998) The "foliage is the fruit" hypothesis: complex adaptations in buffalograss (Buchloe dactyloides). Am Midl Nat 140:252–263
Peakall R, Smouse PE, Huff DR (1995) Evolutionary implications of allozyme and RAPD variation in diploid populations of dioecious buffalograss Buchloe dactyloides. Mol Ecol 4:135–147
Peterson G, Seberg O (1997) Phylogenetic analysis of the Triticeae (Poaceae) based on rpoA sequence data. Mol Phylogenet Evol 7:217–230
Petit RJ, Aguinagalde I, de Beaulieu J-L, Bittkau C, Brewer S, Cheddadi R, Ennos R, Fineschi S, Grivet D, Lascoux M, Mohanty A, Müller-Stark G, Demesure-Musch B, Palmé A, Martín JP, Rendell S, Vendramin GG (2003) Glacial refugia: hotspots but not melting pots of genetic diversity. Science 300:1563–1565
Quinn JA, Mowrey DP, Emanuella SM, Whalley RDB (1994) The “foliage is the fruit” hypothesis: Buchloe dactiloides (Poaceae) and shortgrass prairie of North America. Am J Bot 81:1545:1554
Redingbaugh MG, Jones TA, Zhang Y (2000) Ubiquity of the St chloroplast genome in St-containing Triticeae polyploids. Genome 43:846–52
Reeder JR (1971) Notes on Mexican grasses IX Miscellaneous chromosome numbers. Britannia 23:105–117
Reeder JR, Reeder CG (1963) Notes on Mexican grasses II Cyclostachya, a new dioecious genus. Bull Torrey Bot Club 90:193–201
Reeder JR, Rzedowski J (1965) Notes on Mexican grasses III Buchlomimus, another dioecious genus. Britannia 17:26–33
Roder MS, Korzun V, Wendehake K, Plaschke J, Tixier M, Lorey P, Ganal MW (1998) A microsatellite map of wheat. Genetics 149:2007–2023
Rohlf JF (2000) NTSYS-pc: numerical taxonomy and multivariate analysis system. Exeter Software, Setauket, New York
Sneath PHA, Sokal RR (1973) Numerical taxonomy. Freeman, San Francisco
Soltis DE, Soltis PS (2003) The role of phylogenetics in comparative genetics. Plant Physiol 132:1790–800
Weir BS (1996) Genetic analysis II. Sinauer Publishers, Sunderland, MA
Weising K, Nybon H, Wolff K, Meyer W (1995) DNA fingerprinting in plants and fungi. CRC Press, Boca Raton
Wendel JW (2000) Genome evolution in polyploids. Plant Mol Biol 42:225–249
Wu L, Lin H (1984) Salt tolerance and salt uptake in diploid and polyploid buffalograsses (Buchloë dactyloides). J Plant Nutr 17:1905–1928
Wu J, Konstantin VK, Strauss SH (1998) Abundant mitochondrial genome diversity, population differentiation and convergent evolution in pines. Genetics 150:1605–1614
Yaneshita M, Ohmura T, Sakamura T, Ogihara Y (1993) Phylogenetic relationships of turfgrasses as revealed by restriction fragment analysis of chloroplast DNA. Theor Appl Genet 87:129–135
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by B. Friebe
Rights and permissions
About this article
Cite this article
Budak, H., Shearman, R.C., Gulsen, O. et al. Understanding ploidy complex and geographic origin of the Buchloe dactyloides genome using cytoplasmic and nuclear marker systems. Theor Appl Genet 111, 1545–1552 (2005). https://doi.org/10.1007/s00122-005-0083-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-005-0083-3