Abstract
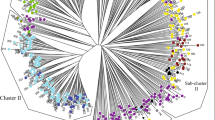
Genetic variations and relationships among cultivated and wild genotypes of five taxa of Indian Luffa were examined using inter simple sequence repeats (ISSR), directed amplification of minisatellite DNA (DAMD) markers and morphological characterization. Morphometric evaluation of 21 discrete characters in 51 representative accessions segregated the five taxa of Luffa in three main clusters: the two wild species (L. echinata, L. graveolens) in the first, and the cultivated L. aegyptiaca (smooth gourd) and L. acutangula (ridged gourd)/L. hermaphrodita (Hermaphrodite luffa) in the second and third clusters, respectively. Cumulative data analysis of 15 ISSR and seven DAMD markers revealed high percentage polymorphism (97.67 %), moderate genetic distance (0.06–0.72, avg. 0.51), and low heterozygosity and Shannon index values (H = 0.15; I = 0.22) across all the 76 genotypes assayed. A UPGMA dendrogram, based on the combined marker data, resolved the five taxa in two main clusters with high bootstrap support. The morphological and molecular trees showed incongruence in the number of main clusters resolved and in the disposition of the wild and cultivated taxa in different sub-clusters. The cluster analyses and PCoA plots revealed a nested grouping of the hermaphrodite luffa within the ridge gourd group. The Bayesian STRUCTURE analysis identified three genetic clusters for the five assumed taxa. Outcrossing test revealed a mixed mating system in Indian Luffa. This is the first ever report on the mating system in Luffa using molecular markers. The study also demonstrates the utility of using more than one DNA marker in the assessment of molecular diversity in a widely cultivated crop genus like Luffa with a narrow genetic base.





Similar content being viewed by others
References
Ahmed B, Alam T, Khan SA (2001) Hepatoprotective activity of Luffa echinata fruits. J Ethnopharmacol 76:187–189
Behera TK, Gaikward AB, Singh AK, Staub JE (2008) Relative efficiency of DNA markers (RAPD, ISSR and AFLP) in detecting genetic diversity of bitter gourd (Momordica charantia L.). J Sci Food Agric 88:733–737
Bhardwaj DR, Kumar V, Lal H, Rai M (2009) Inbreeding depression in sponge gourd [Luffa cylindrica (L.) Roem.]. Veg Sci 36:167–170
Bhattacharyya M, Chakraborty SK (2014) Luffa cylindrica as a host plant for pollinator bees—a study based in West Midnapore, West Bengal, India. J Entomol Zool Stud 2:21–26
Bhawna, Abdin MZ, Arya L, Saha D, Sureja AK, Pandey C, Verma CM (2014) Population structure and genetic diversity in bottle gourd [Lagenaria siceraria (Mol.) Standl.] germplasm from India assessed by ISSR markers. Plant Syst Evol 300:767–773
Chakravarty HL (1982) Cucurbitaceae. Fascicles of Flora of India (Fascicle 11). Botanical Survey of India, Howrah, pp 1–136
Chandra U (1995) Distribution, domestication and genetic diversity of Luffa gourd in Indian sub-continent. Indian J Plant Genet Resour 8:189–196
Chawla R, Thakur P, Chowdhry A, Jaiswal S, Sharma A, Goel R et al (2013) Evidence based herbal drug standardization approach in coping with challenges of holistic management of diabetes: a dreadful lifestyle disorder of 21st century. J Diabetes Metab Disord 12:35
Dongargaon TN, Shashidhar VE, Wali AA, Kulkarni SK (2014) A unique nonsurgical management of internal hemorrhoids by Jimutaka Lepa. Anc Sci Life 33:176–181
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:574–578
Filipowicz N, Schaefer H, Renner SS (2014) Revisiting Luffa (Cucurbitaceae) 25 Years after C. Heiser: species boundaries and application of names tested with plastid and nuclear DNA sequences. Syst Bot 39:205–215
Freitas MLM, Sebbenn AM, Moraes MLT, Lemos EGM (2004) Mating system of a population of Myracrodruon urundeuva F.F. & M.F. Allemão using the fAFLP molecular marker. Genet Mol Biol 27:425–431
Gaiotto FA, Bramucci M, Grattapaglia D (1997) Estimation of outcrossing rate in a breeding population of Eucalyptus urophylla with dominant RAPD and AFLP markers. Theor Appl Genet 95:842–849
Ha H, Lim H, Lee M, Shin I, Jeon WY, Kim J et al (2015) Luffa cylindrica suppresses development of dermatophagoides farinae-induced atopic dermatitis-like skin lesions in Nc/Nga mice. Pharm Biol 53:555–562
Heath DD, Iwama GK, Devlin RH (1993) PCR primed with VNTR core sequences yield species specific patterns and hypervariable probes. Nucleic Acids Res 21:5782–5785
Hedrick PW (1983) Genetics of populations. Science Books International, Boston, MA
Heiser CB, Schilling EE (1988) Phylogeny and distribution of Luffa (Cucurbitaceae). Biotropica 20:185–191
Hu JB, Li JW, Wang LJ, Liu LJ, Si SW (2011) Utilization of a set of high-polymorphism DAMD markers for genetic analysis of a cucumber germplasm collection. Acta Physiol Plant 33:227–231
Idress M, Irshad M (2014) Molecular markers in plants for analysis of genetic diversity: a review. Europ Acad Res 2:1513–1540
Jadhav VB, Thakare VN, Suralkar AA, Naik SR (2013) Ameliorative effect of Luffa acutangula Roxb. on doxorubicin induced cardiac and nephrotoxicity in mice. Ind J Exp Biol 51:149–156
Jayaramappa KV, Pattabhiramaiah M, Bhargava HR (2011) Influence of bee-attractants on yield parameters of ridge gourd (Luffa acutangula L.) (Cucurbitaceae). World Appl Sci J 15:457–462
Jeffrey C (1980) Further notes on Cucurbitaceae: V. The Cucurbitaceae of the Indian subcontinent. Kew Bull 34:789–806
Marr KL, Bhattarai NK, Xia Y (2005a) Allozymic, morphological, and phenological diversity in cultivated collections of Luffa acutangula (Cucurbitaceae) from China, Laos and Nepal and allozyme divergence between L. acutangula and L. aegyptiaca. Econ Bot 59:154–165
Marr KL, Xia Y, Bhattarai NK (2005b) Allozymic, morphological, phenological, linguistic, plant use, and nutritional data on wild and cultivated collections of Luffa aegyptiaca Mill. (Cucurbitaceae) from Nepal, southern China, and northern Laos. Econ Bot 59:137–153
Mensah BA, Kudom AA (2010) Relating bee activity to pollination of Luffa aegyptiaca Mill. in Ghana. J Apic Res 49:192–196
Page RDM (2001) TreeView (Win32), ver. 1.6.5. http://taxonomy.zoology.gla.ac.uk/rod/rod.html
Papanicolaou GC, Psarra E, Anastasiou D (2015) Manufacturing and mechanical response optimization of epoxy resin/Luffa cylindrica composite. J Appl Polym Sci 132:41992
Paris HS, Yonash N, Portnoy V, Mozes-Daube N, Tzuri G, Katzir N (2003) Assessment of genetic relationships in Cucurbita pepo (Cucurbitaceae) using DNA markers. Theor Appl Genet 106:971–978
Pavlicek A, Hrda S, Flegr J (1999) FreeTree–Freeware program for construction of phylogenetic trees on the basis of distance data and bootstrapping/jackknife analysis of the tree robustness—application in the RAPD analysis of the genus Frenkelia. Folia Biol 45:97–99. http://www.natur.cuni.cz/_flegr/free.tree.html
Payel D, Mala P, Sunita S (2015) Inter-genus variation analysis in few members of Cucurbitaceae based on ISSR markers. Biotechnol Biotechnol Equip 29:882–886
Peakall R, Smouse PE (2006) GENAlEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225–238
Prakash K, Pandey A, Radhamani J, Bisht IS (2013) Morphological variability in cultivated and wild species of Luffa (Cucurbitaceae) from India. Genet Resour Crop Evol 60:2319–2329
Prakash K, Pati K, Arya L, Pandey A, Verma M (2014) Population structure and diversity in cultivated and wild Luffa species. Biochem Syst Ecol 56:165–170
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Renner SS, Pandey AK (2013) The Cucurbitaceae of India: accepted names, synonyms, geographic distribution, and information on images and DNA sequences. PhytoKeys 20:53–118
Richharia RH (1948) Sex inheritance in Luffa acutangula. Curr Sci 17:359
Ritland K (1990) A series of FORTRAN computer programs for estimating plant mating systems. J Hered 81:235–237
Ritland K, Jain SA (1981) A model for the estimation of outcrossing rate and gene frequencies using n independent loci. Heredity 47:35–52
Shahidi A, Jalilnejad N, Jalilnejad E (2015) A study on adsorption of cadmium (II) ions from aqueous solution using Luffa cylindrica. Desalin. Water Treat 53:3570–3579
Shang HL, Li CM, Yang ZY, Che DH, Cao JY, Yu Y (2012) Luffa echinata cell (HT-29) death by triggering the mitochondrial apoptosis pathway. Molecules 17:5780–5794
Sharma D, Rawat I, Goel HC (2015) Anticancer and antiinflammatory activity of some dietary cucurbits. Ind J Exp Biol 53:216–221
Sikdar B, Bhattacharya M, Mukherjee A, Banerjee A, Ghosh E, Ghosh B, Roy SC (2010) Genetic diversity in important members of Cucurbitaceae using isozyme, RAPD and ISSR markers. Biol Plant 54:135–140
Singh BP (1991) Interspecific hybridization in between New and Old-world species of Luffa and its phylogenetic implication. Cytologia 56:359–365
Singh PK, Dasgupta SK (2005) Hybrid Loofah. J New Seeds 6:211–215
Singh AK, Kumar S, Singh H, Rai VP, Singh BD, Pandey S (2015) Genetic diversity in Indian snapmelon (Cucumis melo var. momordica) accessions revealed by ISSR markers. Plant Omics 18:9–16
Swofford DL (2002) Phylogenetic analysis using parsimony (*and other methods), ver.4.0b10. Sinauer Associates, Sunderland
Verma VK, Behera TK, Munshi AD, Parida SK, Mohapatra T (2007) Genetic diversity of ash gourd [Benincasa hispida (Thunb.) Cogn.] inbred lines based on RAPD and ISSR markers and their hybrid performance. Sci Hortic 113:231–237
Zietkiewicz E, Rafalski A, Labuda D (1994) Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20:176–183
Acknowledgments
The authors are grateful to the Director, CSIR-National Botanical Research Institute, Lucknow for extending necessary facilities and encouragement. The authors also wish to express their sincere gratitude to the Director, Birbal Sahni Institute of Palaeobotany (BSIP), Lucknow for having permitted them to carry out SEM studies; to Dr. P.E. Rajasekharan, ICAR-Indian Institute of Horticultural Research (ICAR-IIHR), Bangalore; to Dr. T. Pradeepkumar, College of Horticulture, Kerala Agriculture University, Thrissur for having provided seed samples of the standard cultivars and released varieties of Luffa from their germplasm collections; and to Dr. V. Sampath Kumar, the then Indian Botanical Liaison Officer at the Royal Botanic Gardens, Kew, for sending images of a few specimens of Luffa housed in the Kew Herbarium. The financial assistance for conducting research under an In-House Project (OLP 083) of CSIR-NBRI is also gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Misra, S., Srivastava, A.K., Verma, S. et al. Phenetic and genetic diversity in Indian Luffa (Cucurbitaceae) inferred from morphometric, ISSR and DAMD markers. Genet Resour Crop Evol 64, 995–1010 (2017). https://doi.org/10.1007/s10722-016-0420-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-016-0420-1