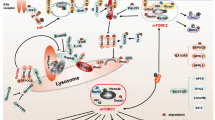
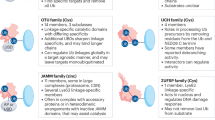
Abstract
Ubiquitination is a reversible cellular process mediated by ubiquitin-conjugating enzymes, whereas deubiquitinating enzymes (DUBs) detach the covalently conjugated ubiquitin from target substrates to counter ubiquitination. DUBs play a crucial role in regulating various signal transduction pathways and biological processes including apoptosis, cell proliferation, DNA damage repair, metastasis, differentiation, etc. Since the transforming growth factor-β (TGF-β) signaling pathway participates in various cellular functions such as inflammation, metastasis and embryogenesis, aberrant regulation of TGF-β signaling induces abnormal cellular functions resulting in numerous diseases. This review focuses on DUBs regulating the TGF-β signaling pathway. We discuss the molecular mechanisms of DUBs involved in TGF-β signaling pathway, and biological and therapeutic implications for various diseases.



Similar content being viewed by others
References
Wilkinson KD (2000) Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Semin Cell Dev Biol 11(3):141–148
Swatek KN, Komander D (2016) Ubiquitin modifications. Cell Res 26(4):399–422
McDowell GS, Philpott A (2013) Non-canonical ubiquitylation: mechanisms and consequences. Int J Biochem Cell Biol 45(8):1833–1842
Grumati P, Dikic I (2018) Ubiquitin signaling and autophagy. J Biol Chem 293(15):5404–5413
Lauwers E, Jacob C, Andre B (2009) K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J Cell Biol 185(3):493–502
Mevissen TET, Komander D (2017) Mechanisms of deubiquitinase specificity and regulation. Annu Rev Biochem 86:159–192
Reyes-Turcu FE, Ventii KH, Wilkinson KD (2009) Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem 78:363–397
Lim KH, Song MH, Baek KH (2016) Decision for cell fate: deubiquitinating enzymes in cell cycle checkpoint. Cell Mol Life Sci 73(7):1439–1455
He M et al (2017) Emerging role of DUBs in tumor metastasis and apoptosis: therapeutic implication. Pharmacol Ther 177:96–107
Burrows JF, Scott CJ, Johnston JA (2010) The DUB/USP17 deubiquitinating enzymes: a gene family within a tandemly repeated sequence, is also embedded within the copy number variable beta-defensin cluster. BMC Genom 11:250
Kim SY et al (2018) PME-1 is regulated by USP36 in ERK and Akt signaling pathways. FEBS Lett 592(9):1575–1588
Kee Y, Huang TT (2016) Role of deubiquitinating enzymes in DNA repair. Mol Cell Biol 36(4):524–544
Nijman SM et al (2005) A genomic and functional inventory of deubiquitinating enzymes. Cell 123(5):773–786
Luise C et al (2011) An atlas of altered expression of deubiquitinating enzymes in human cancer. PLoS One 6(1):e15891
Hu M et al (2002) Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell 111(7):1041–1054
Johnston SC et al (1999) Structural basis for the specificity of ubiquitin C-terminal hydrolases. EMBO J 18(14):3877–3887
Komander D, Clague MJ, Urbe S (2009) Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol 10(8):550–563
Amerik AY, Hochstrasser M (2004) Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta 1695(1–3):189–207
Mao Y et al (2005) Deubiquitinating function of ataxin-3: insights from the solution structure of the Josephin domain. Proc Natl Acad Sci USA 102(36):12700–12705
Nicastro G et al (2005) The solution structure of the Josephin domain of ataxin-3: structural determinants for molecular recognition. Proc Natl Acad Sci USA 102(30):10493–10498
Iyer LM, Koonin EV, Aravind L (2004) Novel predicted peptidases with a potential role in the ubiquitin signaling pathway. Cell Cycle 3(11):1440–1450
Imamura T, Oshima Y, Hikita A (2013) Regulation of TGF-beta family signalling by ubiquitination and deubiquitination. J Biochem 154(6):481–489
Zhang J et al (2014) The regulation of TGF-beta/SMAD signaling by protein deubiquitination. Protein Cell 5(7):503–517
Fraile JM et al (2012) Deubiquitinases in cancer: new functions and therapeutic options. Oncogene 31(19):2373–2388
Derynck R, Zhang YE (2003) Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 425(6958):577–584
Weiss A, Attisano L (2013) The TGFbeta superfamily signaling pathway. Wiley Interdiscip Rev Dev Biol 2(1):47–63
Schmierer B, Hill CS (2007) TGFbeta-SMAD signal transduction: molecular specificity and functional flexibility. Nat Rev Mol Cell Biol 8(12):970–982
Zhang YE (2009) Non-Smad pathways in TGF-beta signaling. Cell Res 19(1):128–139
Conery AR et al (2004) Akt interacts directly with Smad3 to regulate the sensitivity to TGF-beta induced apoptosis. Nat Cell Biol 6(4):366–372
Remy I, Montmarquette A, Michnick SW (2004) PKB/Akt modulates TGF-beta signalling through a direct interaction with Smad3. Nat Cell Biol 6(4):358–365
Yamashita M et al (2008) TRAF6 mediates Smad-independent activation of JNK and p38 by TGF-beta. Mol Cell 31(6):918–924
Ozdamar B et al (2005) Regulation of the polarity protein Par6 by TGFbeta receptors controls epithelial cell plasticity. Science 307(5715):1603–1609
Xu P, Liu J, Derynck R (2012) Post-translational regulation of TGF-beta receptor and Smad signaling. FEBS Lett 586(14):1871–1884
Wang RN et al (2014) Bone morphogenetic protein (BMP) signaling in development and human diseases. Genes Dis 1(1):87–105
Stevenson LF et al (2007) The deubiquitinating enzyme USP2a regulates the p53 pathway by targeting Mdm2. EMBO J 26(4):976–986
Allende-Vega N et al (2010) MdmX is a substrate for the deubiquitinating enzyme USP2a. Oncogene 29(3):432–441
Shan J, Zhao W, Gu W (2009) Suppression of cancer cell growth by promoting cyclin D1 degradation. Mol Cell 36(3):469–476
Shi Y et al (2011) Ubiquitin-specific cysteine protease 2a (USP2a) regulates the stability of aurora-A. J Biol Chem 286(45):38960–38968
Tong X et al (2012) USP2a protein deubiquitinates and stabilizes the circadian protein CRY1 in response to inflammatory signals. J Biol Chem 287(30):25280–25291
Mahul-Mellier AL et al (2012) De-ubiquitinating proteases USP2a and USP2c cause apoptosis by stabilising RIP1. Biochim Biophys Acta 1823(8):1353–1365
Li Y et al (2013) USP2a positively regulates TCR-induced NF-kappaB activation by bridging MALT1-TRAF6. Protein Cell 4(1):62–70
He X et al (2013) USP2a negatively regulates IL-1beta- and virus-induced NF-kappaB activation by deubiquitinating TRAF6. J Mol Cell Biol 5(1):39–47
Kim J et al (2012) The ubiquitin-specific protease USP2a enhances tumor progression by targeting cyclin A1 in bladder cancer. Cell Cycle 11(6):1123–1130
Tao BB et al (2013) Up-regulation of USP2a and FASN in gliomas correlates strongly with glioma grade. J Clin Neurosci 20(5):717–720
Graner E et al (2004) The isopeptidase USP2a regulates the stability of fatty acid synthase in prostate cancer. Cancer Cell 5(3):253–261
Zhao Y et al (2018) USP2a supports metastasis by tuning TGF-beta signaling. Cell Rep 22(9):2442–2454
Clerici M et al (2014) The DUSP-Ubl domain of USP4 enhances its catalytic efficiency by promoting ubiquitin exchange. Nat Commun 5:5399
Li Z et al (2016) USP4 inhibits p53 and NF-kappaB through deubiquitinating and stabilizing HDAC2. Oncogene 35(22):2902–2912
Zhang X et al (2011) USP4 inhibits p53 through deubiquitinating and stabilizing ARF-BP1. EMBO J 30(11):2177–2189
Liu H et al (2015) The deubiquitylating enzyme USP4 cooperates with CtIP in DNA double-strand break end resection. Cell Rep 13(1):93–107
Hou X et al (2013) Ubiquitin-specific protease 4 promotes TNF-alpha-induced apoptosis by deubiquitination of RIP1 in head and neck squamous cell carcinoma. FEBS Lett 587(4):311–316
Xiao N et al (2012) Ubiquitin-specific protease 4 (USP4) targets TRAF2 and TRAF6 for deubiquitination and inhibits TNFalpha-induced cancer cell migration. Biochem J 441(3):979–986
Zhao B et al (2009) The ubiquitin specific protease 4 (USP4) is a new player in the Wnt signalling pathway. J Cell Mol Med 13(8B):1886–1895
Kwon SK, Kim EH, Baek KH (2017) RNPS1 is modulated by ubiquitin-specific protease 4. FEBS Lett 591(2):369–381
Park JK et al (2016) Structural basis for recruiting and shuttling of the spliceosomal deubiquitinase USP4 by SART3. Nucleic Acids Res 44(11):5424–5437
Song EJ et al (2010) The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes Dev 24(13):1434–1447
Lin R et al (2017) USP4 interacts and positively regulates IRF8 function via K48-linked deubiquitination in regulatory T cells. FEBS Lett 591(12):1677–1686
Zhang L et al (2012) USP4 is regulated by AKT phosphorylation and directly deubiquitylates TGF-beta type I receptor. Nat Cell Biol 14(7):717–726
Kavsak P et al (2000) Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell 6(6):1365–1375
Cao WH et al (2016) USP4 promotes invasion of breast cancer cells via Relaxin/TGF-beta1/Smad2/MMP-9 signal. Eur Rev Med Pharmacol Sci 20(6):1115–1122
Xu Y, Yu Q, Liu Y (2018) Serum relaxin-2 as a novel biomarker for prostate cancer. Br J Biomed Sci 75(3):145–148
Ma J et al (2013) Role of relaxin-2 in human primary osteosarcoma. Cancer Cell Int 13(1):59
Mehner C et al (2014) Tumor cell-produced matrix metalloproteinase 9 (MMP-9) drives malignant progression and metastasis of basal-like triple negative breast cancer. Oncotarget 5(9):2736–2749
Al-Hakim AK et al (2008) Control of AMPK-related kinases by USP9X and atypical Lys(29)/Lys(33)-linked polyubiquitin chains. Biochem J 411(2):249–260
Fischer-Vize JA, Rubin GM, Lehmann R (1992) The fat facets gene is required for Drosophila eye and embryo development. Development 116(4):985–1000
Schwickart M et al (2010) Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature 463(7277):103–107
Vong QP et al (2005) Chromosome alignment and segregation regulated by ubiquitination of survivin. Science 310(5753):1499–1504
Engel K et al (2016) USP9X stabilizes XIAP to regulate mitotic cell death and chemoresistance in aggressive B-cell lymphoma. EMBO Mol Med 8(8):851–862
Nagai H et al (2009) Ubiquitin-like sequence in ASK1 plays critical roles in the recognition and stabilization by USP9X and oxidative stress-induced cell death. Mol Cell 36(5):805–818
Huntwork-Rodriguez S et al (2013) JNK-mediated phosphorylation of DLK suppresses its ubiquitination to promote neuronal apoptosis. J Cell Biol 202(5):747–763
Theard D et al (2010) USP9x-mediated deubiquitination of EFA6 regulates de novo tight junction assembly. EMBO J 29(9):1499–1509
Taya S et al (1998) The Ras target AF-6 is a substrate of the fam deubiquitinating enzyme. J Cell Biol 142(4):1053–1062
Mouchantaf R et al (2006) The ubiquitin ligase itch is auto-ubiquitylated in vivo and in vitro but is protected from degradation by interacting with the deubiquitylating enzyme FAM/USP9X. J Biol Chem 281(50):38738–38747
Murray RZ, Jolly LA, Wood SA (2004) The FAM deubiquitylating enzyme localizes to multiple points of protein trafficking in epithelia, where it associates with E-cadherin and beta-catenin. Mol Biol Cell 15(4):1591–1599
Marx C et al (2010) ErbB2 trafficking and degradation associated with K48 and K63 polyubiquitination. Cancer Res 70(9):3709–3717
Murtaza M et al (2015) La FAM fatale: USP9X in development and disease. Cell Mol Life Sci 72(11):2075–2089
Dupont S et al (2009) FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell 136(1):123–135
Xie F et al (2014) Regulation of TGF-beta superfamily signaling by SMAD mono-ubiquitination. Cells 3(4):981–993
Wu Y et al (2017) Aberrant phosphorylation of SMAD4 Thr277-mediated USP9x-SMAD4 interaction by free fatty acids promotes breast cancer metastasis. Cancer Res 77(6):1383–1394
Kinlaw WB et al (2016) Fatty acids and breast cancer: make them on site or have them delivered. J Cell Physiol 231(10):2128–2141
Boden G (2011) Obesity, insulin resistance and free fatty acids. Curr Opin Endocrinol Diabetes Obes 18(2):139–143
Xie Y et al (2013) Deubiquitinase FAM/USP9X interacts with the E3 ubiquitin ligase SMURF1 protein and protects it from ligase activity-dependent self-degradation. J Biol Chem 288(5):2976–2985
Stegeman S et al (2013) Loss of Usp9x disrupts cortical architecture, hippocampal development and TGFbeta-mediated axonogenesis. PLoS One 8(7):e68287
Harper S et al (2014) Structure and catalytic regulatory function of ubiquitin specific protease 11N-terminal and ubiquitin-like domains. Biochemistry 53(18):2966–2978
Wu HC et al (2014) USP11 regulates PML stability to control Notch-induced malignancy in brain tumours. Nat Commun 5:3214
Lee EW et al (2015) USP11-dependent selective cIAP2 deubiquitylation and stabilization determine sensitivity to Smac mimetics. Cell Death Differ 22(9):1463–1476
Zhou Z et al (2017) Regulation of XIAP turnover reveals a role for USP11 in promotion of tumorigenesis. EBioMedicine 15:48–61
Kapadia B et al (2018) Fatty acid synthase induced S6Kinase facilitates USP11-eIF4B complex formation for sustained oncogenic translation in DLBCL. Nat Commun 9(1):829
Wang D et al (2018) Phosphorylated E2F1 is stabilized by nuclear USP11 to drive Peg10 gene expression and activate lung epithelial cells. J Mol Cell Biol 10(1):60–73
Zhang E et al (2016) Ubiquitin-specific protease 11 (USP11) functions as a tumor suppressor through deubiquitinating and stabilizing VGLL4 protein. Am J Cancer Res 6(12):2901–2909
Deng T et al (2018) Deubiquitylation and stabilization of p21 by USP11 is critical for cell-cycle progression and DNA damage responses. Proc Natl Acad Sci USA 115(18):4678–4683
Schoenfeld AR et al (2004) BRCA2 is ubiquitinated in vivo and interacts with USP11, a deubiquitinating enzyme that exhibits prosurvival function in the cellular response to DNA damage. Mol Cell Biol 24(17):7444–7455
Wiltshire TD et al (2010) Sensitivity to poly(ADP-ribose) polymerase (PARP) inhibition identifies ubiquitin-specific peptidase 11 (USP11) as a regulator of DNA double-strand break repair. J Biol Chem 285(19):14565–14571
Yu M et al (2016) USP11 is a negative regulator to gammaH2AX ubiquitylation by RNF8/RNF168. J Biol Chem 291(2):959–967
Shah P et al (2017) Regulation of XPC deubiquitination by USP11 in repair of UV-induced DNA damage. Oncotarget 8(57):96522–96535
Yamaguchi T et al (2007) The deubiquitinating enzyme USP11 controls an IkappaB kinase alpha (IKKalpha)-p53 signaling pathway in response to tumor necrosis factor alpha (TNFalpha). J Biol Chem 282(47):33943–33948
Sun W et al (2010) USP11 negatively regulates TNFalpha-induced NF-kappaB activation by targeting on IkappaBalpha. Cell Signal 22(3):386–394
Lim KH et al (2016) Ubiquitin-specific protease 11 functions as a tumor suppressor by modulating Mgl-1 protein to regulate cancer cell growth. Oncotarget 7(12):14441–14457
Ideguchi H et al (2002) Structural and functional characterization of the USP11 deubiquitinating enzyme, which interacts with the RanGTP-associated protein RanBPM. Biochem J 367(Pt 1):87–95
Lin CH, Chang HS, Yu WC (2008) USP11 stabilizes HPV-16E7 and further modulates the E7 biological activity. J Biol Chem 283(23):15681–15688
Ke JY et al (2014) USP11 regulates p53 stability by deubiquitinating p53. J Zhejiang Univ Sci B 15(12):1032–1038
Al-Salihi MA et al (2012) USP11 augments TGFbeta signalling by deubiquitylating ALK5. Open Biol 2(6):120063
Jacko AM et al (2016) De-ubiquitinating enzyme, USP11, promotes transforming growth factor beta-1 signaling through stabilization of transforming growth factor beta receptor II. Cell Death Dis 7(11):e2474
Li J, Wang G, Sun X (2014) Transforming growth factor beta regulates beta-catenin expression in lung fibroblast through NF-kappaB dependent pathway. Int J Mol Med 34(5):1219–1224
Garcia DA et al (2018) USP11 enhances TGFbeta-induced epithelial-mesenchymal plasticity and human breast cancer metastasis. Mol Cancer Res 16(7):1172–1184
Ward SJ et al (2018) The structure of the deubiquitinase USP15 reveals a misaligned catalytic triad and an open ubiquitin-binding channel. J Biol Chem [Epub ahead of print]
Schweitzer K et al (2007) CSN controls NF-kappaB by deubiquitinylation of IkappaBalpha. EMBO J 26(6):1532–1541
Kawahara K et al (2000) Down-regulation of beta-catenin by the colorectal tumor suppressor APC requires association with Axin and beta-catenin. J Biol Chem 275(12):8369–8374
Huang X et al (2009) The COP9 signalosome mediates beta-catenin degradation by deneddylation and blocks adenomatous polyposis coli destruction via USP15. J Mol Biol 391(4):691–702
Zou Q et al (2014) USP15 stabilizes MDM2 to mediate cancer-cell survival and inhibit antitumor T cell responses. Nat Immunol 15(6):562–570
Vos RM et al (2009) The ubiquitin-specific peptidase USP15 regulates human papillomavirus type 16 E6 protein stability. J Virol 83(17):8885–8892
Inui M et al (2011) USP15 is a deubiquitylating enzyme for receptor-activated SMADs. Nat Cell Biol 13(11):1368–1375
Eichhorn PJ et al (2012) USP15 stabilizes TGF-beta receptor I and promotes oncogenesis through the activation of TGF-beta signaling in glioblastoma. Nat Med 18(3):429–435
Iyengar PV et al (2015) USP15 regulates SMURF2 kinetics through C-lobe mediated deubiquitination. Sci Rep 5:14733
Liu WT et al (2017) TGF-beta upregulates the translation of USP15 via the PI3K/AKT pathway to promote p53 stability. Oncogene 36(19):2715–2723
Lam YA et al (1997) Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature 385(6618):737–740
Peth A et al (2013) Ubiquitinated proteins activate the proteasomal ATPases by binding to Usp14 or Uch37 homologs. J Biol Chem 288(11):7781–7790
Chen Y et al (2012) Expression and clinical significance of UCH37 in human esophageal squamous cell carcinoma. Dig Dis Sci 57(9):2310–2317
Wang L et al (2014) High expression of UCH37 is significantly associated with poor prognosis in human epithelial ovarian cancer. Tumour Biol 35(11):11427–11433
Zhou Z et al (2018) The deubiquitinase UCHL5/UCH37 positively regulates Hedgehog signaling by deubiquitinating smoothened. J Mol Cell Biol 10(3):243–257
Mahanic CS et al (2015) Regulation of E2 promoter binding factor 1 (E2F1) transcriptional activity through a deubiquitinating enzyme, UCH37. J Biol Chem 290(44):26508–26522
Randles L et al (2016) The proteasome ubiquitin receptor hRpn13 and its interacting deubiquitinating enzyme Uch37 are required for proper cell cycle progression. J Biol Chem 291(16):8773–8783
Wicks SJ et al (2005) The deubiquitinating enzyme UCH37 interacts with Smads and regulates TGF-beta signalling. Oncogene 24(54):8080–8084
Cutts AJ et al (2011) Early phase TGFbeta receptor signalling dynamics stabilised by the deubiquitinase UCH37 promotes cell migratory responses. Int J Biochem Cell Biol 43(4):604–612
Edelmann MJ et al (2009) Structural basis and specificity of human otubain 1-mediated deubiquitination. Biochem J 418(2):379–390
Zhou Y et al (2014) OTUB1 promotes metastasis and serves as a marker of poor prognosis in colorectal cancer. Mol Cancer 13:258
Iglesias-Gato D et al (2015) OTUB1 de-ubiquitinating enzyme promotes prostate cancer cell invasion in vitro and tumorigenesis in vivo. Mol Cancer 14:8
Iglesias-Gato D et al (2015) Erratum: OTUB1 de-ubiquitinating enzyme promotes prostate cancer cell invasion in vitro and tumorigenesis in vivo. Mol Cancer 14:88
Wang Y et al (2016) OTUB1-catalyzed deubiquitination of FOXM1 facilitates tumor progression and predicts a poor prognosis in ovarian cancer. Oncotarget 7(24):36681–36697
Wang YQ et al (2016) Upregulation of the non-coding RNA OTUB1-isoform 2 contributes to gastric cancer cell proliferation and invasion and predicts poor gastric cancer prognosis. Int J Biol Sci 12(5):545–557
Weng W et al (2016) OTUB1 promotes tumor invasion and predicts a poor prognosis in gastric adenocarcinoma. Am J Transl Res 8(5):2234–2244
Ni Q et al (2017) Expression of OTUB1 in hepatocellular carcinoma and its effects on HCC cell migration and invasion. Acta Biochim Biophys Sin (Shanghai) 49(8):680–688
Zhou H et al (2018) OTUB1 promotes esophageal squamous cell carcinoma metastasis through modulating Snail stability. Oncogene 37(25):3356–3368
Karunarathna U et al (2016) OTUB1 inhibits the ubiquitination and degradation of FOXM1 in breast cancer and epirubicin resistance. Oncogene 35(11):1433–1444
Li S et al (2010) Regulation of virus-triggered signaling by OTUB1- and OTUB2-mediated deubiquitination of TRAF3 and TRAF6. J Biol Chem 285(7):4291–4297
Zhao L et al (2018) OTUB1 protein suppresses mTOR complex 1 (mTORC1) activity by deubiquitinating the mTORC1 inhibitor DEPTOR. J Biol Chem 293(13):4883–4892
Li Y et al (2014) Monoubiquitination is critical for ovarian tumor domain-containing ubiquitin aldehyde binding protein 1 (Otub1) to suppress UbcH5 enzyme and stabilize p53 protein. J Biol Chem 289(8):5097–5108
Goncharov T et al (2013) OTUB1 modulates c-IAP1 stability to regulate signalling pathways. EMBO J 32(8):1103–1114
Blackford AN, Stewart GS (2011) When cleavage is not attractive: non-catalytic inhibition of ubiquitin chains at DNA double-strand breaks by OTUB1. DNA Repair (Amst) 10(2):245–249
Chen Y et al (2017) Otub1 stabilizes MDMX and promotes its proapoptotic function at the mitochondria. Oncotarget 8(7):11053–11062
Herhaus L et al (2013) OTUB1 enhances TGFbeta signalling by inhibiting the ubiquitylation and degradation of active SMAD2/3. Nat Commun 4:2519
Sato Y et al (2008) Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature 455(7211):358–362
Davies CW et al (2011) Structural and thermodynamic comparison of the catalytic domain of AMSH and AMSH-LP: nearly identical fold but different stability. J Mol Biol 413(2):416–429
Ibarrola N et al (2004) Cloning of a novel signaling molecule, AMSH-2, that potentiates transforming growth factor beta signaling. BMC Cell Biol 5:2
Fan YH et al (2011) USP4 targets TAK1 to downregulate TNFalpha-induced NF-kappaB activation. Cell Death Differ 18(10):1547–1560
Wang PJ et al (2001) An abundance of X-linked genes expressed in spermatogonia. Nat Genet 27(4):422–426
Ribarski I et al (2009) USP26 gene variations in fertile and infertile men. Hum Reprod 24(2):477–484
Ma Q et al (2016) A novel missense mutation in USP26 gene is associated with nonobstructive azoospermia. Reprod Sci 23(10):1434–1441
Dirac AM, Bernards R (2010) The deubiquitinating enzyme USP26 is a regulator of androgen receptor signaling. Mol Cancer Res 8(6):844–854
Zhang W et al (2015) Evidence from enzymatic and meta-analyses does not support a direct association between USP26 gene variants and male infertility. Andrology 3(2):271–279
Typas D et al (2015) The de-ubiquitylating enzymes USP26 and USP37 regulate homologous recombination by counteracting RAP80. Nucleic Acids Res 43(14):6919–6933
Ning B et al (2017) USP26 functions as a negative regulator of cellular reprogramming by stabilising PRC1 complex components. Nat Commun 8(1):349
Lahav-Baratz S et al (2017) The testis-specific USP26 is a deubiquitinating enzyme of the ubiquitin ligase Mdm2. Biochem Biophys Res Commun 482(1):106–111
Kit Leng Lui S et al (2017) USP26 regulates TGF-beta signaling by deubiquitinating and stabilizing SMAD7. EMBO Rep 18(5):797–808
Komander D et al (2008) The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Mol Cell 29(4):451–464
Brummelkamp TR et al (2003) Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature 424(6950):797–801
Kovalenko A et al (2003) The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature 424(6950):801–805
Trompouki E et al (2003) CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature 424(6950):793–796
Yoshida H et al (2005) The tumor suppressor cylindromatosis (CYLD) acts as a negative regulator for toll-like receptor 2 signaling via negative cross-talk with TRAF6 AND TRAF7. J Biol Chem 280(49):41111–41121
Lim JH et al (2007) Tumor suppressor CYLD regulates acute lung injury in lethal Streptococcus pneumoniae infections. Immunity 27(2):349–360
Chen W et al (2003) Conversion of peripheral CD4+ CD25− naive T cells to CD4+ CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med 198(12):1875–1886
Zhao Y et al (2011) The deubiquitinase CYLD targets Smad7 protein to regulate transforming growth factor beta (TGF-beta) signaling and the development of regulatory T cells. J Biol Chem 286(47):40520–40530
Komander D, Barford D (2008) Structure of the A20 OTU domain and mechanistic insights into deubiquitination. Biochem J 409(1):77–85
Shembade N, Harhaj E (2010) A20 inhibition of NFkappaB and inflammation: targeting E2:E3 ubiquitin enzyme complexes. Cell Cycle 9(13):2481–2482
De Valck D et al (1999) The zinc finger protein A20 interacts with a novel anti-apoptotic protein which is cleaved by specific caspases. Oncogene 18(29):4182–4190
Daniel S et al (2004) A20 protects endothelial cells from TNF-, Fas-, and NK-mediated cell death by inhibiting caspase 8 activation. Blood 104(8):2376–2384
Hovelmeyer N et al (2011) A20 deficiency in B cells enhances B-cell proliferation and results in the development of autoantibodies. Eur J Immunol 41(3):595–601
Chu Y et al (2011) B cells lacking the tumor suppressor TNFAIP3/A20 display impaired differentiation and hyperactivation and cause inflammation and autoimmunity in aged mice. Blood 117(7):2227–2236
Shi CS, Kehrl JH (2010) TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal 3(123):ra42
Inomata M et al (2012) Regulation of toll-like receptor signaling by NDP52-mediated selective autophagy is normally inactivated by A20. Cell Mol Life Sci 69(6):963–979
Jung SM et al (2013) Smad6 inhibits non-canonical TGF-beta1 signalling by recruiting the deubiquitinase A20 to TRAF6. Nat Commun 4:2562
Iyengar PV (2017) Regulation of ubiquitin enzymes in the TGF-beta pathway. Int J Mol Sci 18(4):877
Davis MI et al (2016) Small molecule inhibition of the ubiquitin-specific protease USP2 accelerates cyclin D1 degradation and leads to cell cycle arrest in colorectal cancer and mantle cell lymphoma models. J Biol Chem 291(47):24628–24640
Burkhart RA et al (2013) Mitoxantrone targets human ubiquitin-specific peptidase 11 (USP11) and is a potent inhibitor of pancreatic cancer cell survival. Mol Cancer Res 11(8):901–911
Ernst A et al (2013) A strategy for modulation of enzymes in the ubiquitin system. Science 339(6119):590–595
Tomala MD et al (2018) Identification of small-molecule inhibitors of USP2a. Eur J Med Chem 150:261–267
Ndubaku C, Tsui V (2015) Inhibiting the deubiquitinating enzymes (DUBs). J Med Chem 58(4):1581–1595
Kang JS et al (2008) The type I TGF-beta receptor is covalently modified and regulated by sumoylation. Nat Cell Biol 10(6):654–664
Lee PS et al (2003) Sumoylation of Smad4, the common Smad mediator of transforming growth factor-beta family signaling. J Biol Chem 278(30):27853–27863
Inoue Y, Imamura T (2008) Regulation of TGF-beta family signaling by E3 ubiquitin ligases. Cancer Sci 99(11):2107–2112
Zhang Y et al (2001) Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc Natl Acad Sci USA 98(3):974–979
Lin X, Liang M, Feng XH (2000) Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-beta signaling. J Biol Chem 275(47):36818–36822
Acknowledgements
We would like to thank members of Baek’s laboratory for their critical comments on the manuscript. This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (201600490003).
Author information
Authors and Affiliations
Contributions
SYK: manuscript writing; KHB: manuscript writing, final approval of manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Kim, SY., Baek, KH. TGF-β signaling pathway mediated by deubiquitinating enzymes. Cell. Mol. Life Sci. 76, 653–665 (2019). https://doi.org/10.1007/s00018-018-2949-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-018-2949-y