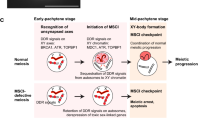
Abstract
Sex chromosome inactivation in male germ cells is a paradigm of epigenetic programming during sexual reproduction. Recent progress has revealed the underlying mechanisms of sex chromosome inactivation in male meiosis. The trigger of chromosome-wide silencing is activation of the DNA damage response (DDR) pathway, which is centered on the mediator of DNA damage checkpoint 1 (MDC1), a binding partner of phosphorylated histone H2AX (γH2AX). This DDR pathway shares features with the somatic DDR pathway recognizing DNA replication stress in the S phase. Additionally, it is likely to be distinct from the DDR pathway that recognizes meiosis-specific double-strand breaks. This review article extensively discusses the underlying mechanism of sex chromosome inactivation.



Similar content being viewed by others
References
Ohno S (1967) Sex chromosomes and sex-linked genes. Springer, Berlin
Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L et al (2003) The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423:825–837
Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K et al (2005) The DNA sequence of the human X chromosome. Nature 434:325–337
Payer B, Lee JT, Namekawa SH (2011) X-inactivation and X-reactivation: epigenetic hallmarks of mammalian reproduction and pluripotent stem cells. Hum Genet 130:265–280
Turner JM (2007) Meiotic sex chromosome inactivation. Development 134:1823–1831
Burgoyne PS, Mahadevaiah SK, Turner JM (2009) The consequences of asynapsis for mammalian meiosis. Nat Rev Genet 10:207–216
Yan W, McCarrey JR (2009) Sex chromosome inactivation in the male. Epigenetics 4:452–456
Inagaki A, Schoenmakers S, Baarends WM (2010) DNA double strand break repair, chromosome synapsis and transcriptional silencing in meiosis. Epigenetics 5:255–266
Cloutier JM, Turner JM (2010) Meiotic sex chromosome inactivation. Curr Biol 20:R962–R963
Heard E, Turner J (2011) Function of the sex chromosomes in mammalian fertility. Cold Spring Harb Perspect Biol 3:a002675
Namekawa SH, Park PJ, Zhang LF, Shima JE, McCarrey JR et al (2006) Postmeiotic sex chromatin in the male germline of mice. Curr Biol 16:660–667
Fallahi M, Getun IV, Wu ZK, Bois PR (2010) A global expression switch marks pachytene initiation during mouse male meiosis. Genes 1:469–483
Song R, Ro S, Michaels JD, Park C, McCarrey JR et al (2009) Many X-linked microRNAs escape meiotic sex chromosome inactivation. Nat Genet 41:488–493
Anguera MC, Ma W, Clift D, Namekawa S, Kelleher RJ 3rd et al (2011) Tsx produces a long noncoding RNA and has general functions in the germline, stem cells, and brain. PLoS Genet 7:e1002248
Turner JM, Mahadevaiah SK, Ellis PJ, Mitchell MJ, Burgoyne PS (2006) Pachytene asynapsis drives meiotic sex chromosome inactivation and leads to substantial postmeiotic repression in spermatids. Dev Cell 10:521–529
Greaves IK, Rangasamy D, Devoy M, Marshall Graves JA, Tremethick DJ (2006) The X and Y chromosomes assemble into H2A.Z-containing [corrected] facultative heterochromatin [corrected] following meiosis. Mol Cell Biol 26:5394–5405
Mueller JL, Mahadevaiah SK, Park PJ, Warburton PE, Page DC et al (2008) The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nat Genet 40:794–799
Fernandez-Capetillo O, Mahadevaiah SK, Celeste A, Romanienko PJ, Camerini-Otero RD et al (2003) H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev Cell 4:497–508
Ichijima Y, Ichijima M, Lou Z, Nussenzweig A, Camerini-Otero RD et al (2011) MDC1 directs chromosome-wide silencing of the sex chromosomes in male germ cells. Genes Dev 25:959–971
Baarends WM, Wassenaar E, van der Laan R, Hoogerbrugge J, Sleddens-Linkels E et al (2005) Silencing of unpaired chromatin and histone H2A ubiquitination in mammalian meiosis. Mol Cell Biol 25:1041–1053
Turner JM, Mahadevaiah SK, Fernandez-Capetillo O, Nussenzweig A, Xu X et al (2005) Silencing of unsynapsed meiotic chromosomes in the mouse. Nat Genet 37:41–47
Baumann C, Daly CM, McDonnell SM, Viveiros MM, De La Fuente R (2011) Chromatin configuration and epigenetic landscape at the sex chromosome bivalent during equine spermatogenesis. Chromosoma 120:227–244
Namekawa SH, VandeBerg JL, McCarrey JR, Lee JT (2007) Sex chromosome silencing in the marsupial male germ line. Proc Natl Acad Sci USA 104:9730–9735
van der Heijden GW, Derijck AA, Posfai E, Giele M, Pelczar P et al (2007) Chromosome-wide nucleosome replacement and H3.3 incorporation during mammalian meiotic sex chromosome inactivation. Nat Genet 39:251–258
Handel MA, Schimenti JC (2010) Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat Rev Genet 11:124–136
Baudat F, Manova K, Yuen JP, Jasin M, Keeney S (2000) Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol Cell 6:989–998
Romanienko PJ, Camerini-Otero RD (2000) The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell 6:975–987
Mahadevaiah SK, Turner JM, Baudat F, Rogakou EP, de Boer P et al (2001) Recombinational DNA double-strand breaks in mice precede synapsis. Nat Genet 27:271–276
Turner JM, Aprelikova O, Xu X, Wang R, Kim S et al (2004) BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. Curr Biol 14:2135–2142
Perera D, Perez-Hidalgo L, Moens PB, Reini K, Lakin N et al (2004) TopBP1 and ATR colocalization at meiotic chromosomes: role of TopBP1/Cut5 in the meiotic recombination checkpoint. Mol Biol Cell 15:1568–1579
Reini K, Uitto L, Perera D, Moens PB, Freire R et al (2004) TopBP1 localises to centrosomes in mitosis and to chromosome cores in meiosis. Chromosoma 112:323–330
Lee AC, Fernandez-Capetillo O, Pisupati V, Jackson SP, Nussenzweig A (2005) Specific association of mouse MDC1/NFBD1 with NBS1 at sites of DNA-damage. Cell Cycle 4:177–182
Chen J, Silver DP, Walpita D, Cantor SB, Gazdar AF et al (1998) Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol Cell 2:317–328
Huen MS, Sy SM, Chen J (2010) BRCA1 and its toolbox for the maintenance of genome integrity. Natl Rev Mol Cell Biol 11:138–148
Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ (2003) MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421:961–966
Lou Z, Minter-Dykhouse K, Wu X, Chen J (2003) MDC1 is coupled to activated CHK2 in mammalian DNA damage response pathways. Nature 421:957–961
Goldberg M, Stucki M, Falck J, D’Amours D, Rahman D et al (2003) MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature 421:952–956
Jungmichel S, Stucki M (2010) MDC1: the art of keeping things in focus. Chromosoma 119:337–349
Ahmed EA, van der Vaart A, Barten A, Kal HB, Chen J et al (2007) Differences in DNA double strand breaks repair in male germ cell types: lessons learned from a differential expression of Mdc1 and 53BP1. DNA Repair (Amst) 6:1243–1254
Bellani MA, Romanienko PJ, Cairatti DA, Camerini-Otero RD (2005) SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm−/− spermatocytes. J Cell Sci 118:3233–3245
Polo SE, Jackson SP (2011) Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev 25:409–433
Cimprich KA, Cortez D (2008) ATR: an essential regulator of genome integrity. Natl Rev Mol Cell Biol 9:616–627
Deans AJ, West SC (2011) DNA interstrand crosslink repair and cancer. Nat Rev Cancer 11:467–480
O’Sullivan RJ, Karlseder J (2010) Telomeres: protecting chromosomes against genome instability. Natl Rev Mol Cell Biol 11:171–181
Shiotani B, Zou L (2009) Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol Cell 33:547–558
Parrilla-Castellar ER, Arlander SJ, Karnitz L (2004) Dial 9–1-1 for DNA damage: the Rad9-Hus1-Rad1 (9–1-1) clamp complex. DNA Repair (Amst) 3:1009–1014
Freire R, Murguia JR, Tarsounas M, Lowndes NF, Moens PB et al (1998) Human and mouse homologs of Schizosaccharomyces pombe rad1(+) and Saccharomyces cerevisiae RAD17: linkage to checkpoint control and mammalian meiosis. Genes Dev 12:2560–2573
Zou L, Elledge SJ (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300:1542–1548
Plug AW, Peters AH, Keegan KS, Hoekstra MF, de Boer P et al (1998) Changes in protein composition of meiotic nodules during mammalian meiosis. J Cell Sci 111(Pt 4):413–423
Burgoyne PS, Mahadevaiah SK, Turner JM (2007) The management of DNA double-strand breaks in mitotic G2, and in mammalian meiosis viewed from a mitotic G2 perspective. Bioessays 29:974–986
Collis SJ, Ciccia A, Deans AJ, Horejsi Z, Martin JS et al (2008) FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the Fanconi anemia core complex. Mol Cell 32:313–324
Huang M, Kim JM, Shiotani B, Yang K, Zou L et al (2010) The FANCM/FAAP24 complex is required for the DNA interstrand crosslink-induced checkpoint response. Mol Cell 39:259–268
Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C et al (2001) Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell 7:249–262
Holloway JK, Mohan S, Balmus G, Sun X, Modzelewski A et al (2011) Mammalian BTBD12 (SLX4) protects against genomic instability during mammalian spermatogenesis. PLoS Genet 7:e1002094
Scully R, Chen J, Ochs RL, Keegan K, Hoekstra M et al (1997) Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell 90:425–435
Wojtasz L, Daniel K, Roig I, Bolcun-Filas E, Xu H et al (2009) Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet 5:e1000702
Daniel K, Lange J, Hached K, Fu J, Anastassiadis K et al (2011) Meiotic homologue alignment and its quality surveillance are controlled by mouse HORMAD1. Nat Cell Biol 13:599–610
Kouznetsova A, Wang H, Bellani M, Camerini-Otero RD, Jessberger R et al (2009) BRCA1-mediated chromatin silencing is limited to oocytes with a small number of asynapsed chromosomes. J Cell Sci 122:2446–2452
An JY, Kim EA, Jiang Y, Zakrzewska A, Kim DE et al (2010) UBR2 mediates transcriptional silencing during spermatogenesis via histone ubiquitination. Proc Natl Acad Sci USA 107:1912–1917
Ashley T, Gaeth AP, Creemers LB, Hack AM, de Rooij DG (2004) Correlation of meiotic events in testis sections and microspreads of mouse spermatocytes relative to the mid-pachytene checkpoint. Chromosoma 113:126–136
Moens PB, Kolas NK, Tarsounas M, Marcon E, Cohen PE et al (2002) The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA–DNA interactions without reciprocal recombination. J Cell Sci 115:1611–1622
Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA et al (2006) MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell 21:187–200
Wang J, Gong Z, Chen J (2011) MDC1 collaborates with TopBP1 in DNA replication checkpoint control. J Cell Biol 193:267–273
Huen MS, Grant R, Manke I, Minn K, Yu X et al (2007) RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131:901–914
Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R et al (2007) Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318:1637–1640
Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J et al (2007) RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131:887–900
Lu LY, Wu J, Ye L, Gavrilina GB, Saunders TL et al (2010) RNF8-dependent histone modifications regulate nucleosome removal during spermatogenesis. Dev Cell 18:371–384
Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA (2010) ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141:970–981
Mahadevaiah SK, Bourc’his D, de Rooij DG, Bestor TH, Turner JM et al (2008) Extensive meiotic asynapsis in mice antagonises meiotic silencing of unsynapsed chromatin and consequently disrupts meiotic sex chromosome inactivation. J Cell Biol 182:263–276
Haaf T, Golub EI, Reddy G, Radding CM, Ward DC (1995) Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc Natl Acad Sci USA 92:2298–2302
Tarsounas M, Davies D, West SC (2003) BRCA2-dependent and independent formation of RAD51 nuclear foci. Oncogene 22:1115–1123
Long DT, Raschle M, Joukov V, Walter JC (2011) Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science 333:84–87
Hu Y, Scully R, Sobhian B, Xie A, Shestakova E et al (2011) RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes Dev 25:685–700
Eijpe M, Offenberg H, Goedecke W, Heyting C (2000) Localisation of RAD50 and MRE11 in spermatocyte nuclei of mouse and rat. Chromosoma 109:123–132
Rogers RS, Inselman A, Handel MA, Matunis MJ (2004) SUMO modified proteins localize to the XY body of pachytene spermatocytes. Chromosoma 113:233–243
Xu X, Aprelikova O, Moens P, Deng CX, Furth PA (2003) Impaired meiotic DNA-damage repair and lack of crossing-over during spermatogenesis in BRCA1 full-length isoform deficient mice. Development 130:2001–2012
Sharan SK, Pyle A, Coppola V, Babus J, Swaminathan S et al (2004) BRCA2 deficiency in mice leads to meiotic impairment and infertility. Development 131:131–142
Holloway JK, Morelli MA, Borst PL, Cohen PE (2010) Mammalian BLM helicase is critical for integrating multiple pathways of meiotic recombination. J Cell Biol 188:779–789
Liang Y, Gao H, Lin SY, Peng G, Huang X et al (2010) BRIT1/MCPH1 is essential for mitotic and meiotic recombination DNA repair and maintaining genomic stability in mice. PLoS Genet 6:e1000826
Goedecke W, Eijpe M, Offenberg HH, van Aalderen M, Heyting C (1999) Mre11 and Ku70 interact in somatic cells, but are differentially expressed in early meiosis. Nat Genet 23:194–198
Ward IM, Minn K, van Deursen J, Chen J (2003) p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol Cell Biol 23:2556–2563
Baarends WM, Wassenaar E, Hoogerbrugge JW, van Cappellen G, Roest HP et al (2003) Loss of HR6B ubiquitin-conjugating activity results in damaged synaptonemal complex structure and increased crossing-over frequency during the male meiotic prophase. Mol Cell Biol 23:1151–1162
van der Laan R, Uringa EJ, Wassenaar E, Hoogerbrugge JW, Sleddens E et al (2004) Ubiquitin ligase Rad18Sc localizes to the XY body and to other chromosomal regions that are unpaired and transcriptionally silenced during male meiotic prophase. J Cell Sci 117:5023–5033
Inagaki A, Sleddens-Linkels E, Wassenaar E, Ooms M, van Cappellen WA et al (2011) Meiotic functions of RAD18. J Cell Sci 124:2837–2850
Baarends WM, Wassenaar E, Hoogerbrugge JW, Schoenmakers S, Sun ZW et al (2007) Increased phosphorylation and dimethylation of XY body histones in the Hr6b-knockout mouse is associated with derepression of the X chromosome. J Cell Sci 120:1841–1851
Barchi M, Mahadevaiah S, Di Giacomo M, Baudat F, de Rooij DG et al (2005) Surveillance of different recombination defects in mouse spermatocytes yields distinct responses despite elimination at an identical developmental stage. Mol Cell Biol 25:7203–7215
Lange J, Pan J, Cole F, Thelen MP, Jasin M et al (2011) ATM controls meiotic double-strand-break formation. Nature 479:237–240
Manterola M, Page J, Vasco C, Berrios S, Parra MT et al (2009) A high incidence of meiotic silencing of unsynapsed chromatin is not associated with substantial pachytene loss in heterozygous male mice carrying multiple simple robertsonian translocations. PLoS Genet 5:e1000625
Ohno S, Cattanach BM (1962) Cytological study of an X-autosome translocation in Mus musculus. Cytogenetics 1:129–140
Homolka D, Ivanek R, Capkova J, Jansa P, Forejt J (2007) Chromosomal rearrangement interferes with meiotic X chromosome inactivation. Genome Res 17:1431–1437
Homolka D, Jansa P, Forejt J (2012) Genetically enhanced asynapsis of autosomal chromatin promotes transcriptional dysregulation and meiotic failure. Chromosoma 121:91–104
Xu Y, Ashley T, Brainerd EE, Bronson RT, Meyn MS et al (1996) Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev 10:2411–2422
MacQueen AJ, Hochwagen A (2011) Checkpoint mechanisms: the puppet masters of meiotic prophase. Trends Cell Biol 21:393–400
San-Segundo PA, Roeder GS (1999) Pch2 links chromatin silencing to meiotic checkpoint control. Cell 97:313–324
Bhalla N, Dernburg AF (2005) A conserved checkpoint monitors meiotic chromosome synapsis in Caenorhabditis elegans. Science 310:1683–1686
Joyce EF, McKim KS (2009) Drosophila PCH2 is required for a pachytene checkpoint that monitors double-strand-break-independent events leading to meiotic crossover formation. Genetics 181:39–51
Li XC, Schimenti JC (2007) Mouse pachytene checkpoint 2 (trip13) is required for completing meiotic recombination but not synapsis. PLoS Genet 3:e130
Roig I, Dowdle JA, Toth A, de Rooij DG, Jasin M, et al. (2010) Mouse TRIP13/PCH2 is required for recombination and normal higher-order chromosome structure during meiosis. PLoS Genet 66(8): e1001062
Li XC, Barringer BC, Barbash DA (2009) The pachytene checkpoint and its relationship to evolutionary patterns of polyploidization and hybrid sterility. Heredity 102:24–30
Royo H, Polikiewicz G, Mahadevaiah SK, Prosser H, Mitchell M et al (2010) Evidence that meiotic sex chromosome inactivation is essential for male fertility. Curr Biol 20:2117–2123
Vernet N, Mahadevaiah SK, Ojarikre OA, Longepied G, Prosser HM et al (2011) The Y-encoded gene zfy2 acts to remove cells with unpaired chromosomes at the first meiotic metaphase in male mice. Curr Biol 21:787–793
Toth A, Jessberger R (2010) Male meiosis: Y keep it silenced? Curr Biol 20:R1022–R1024
Cowell IG, Aucott R, Mahadevaiah SK, Burgoyne PS, Huskisson N et al (2002) Heterochromatin, HP1 and methylation at lysine 9 of histone H3 in animals. Chromosoma 111:22–36
Khalil AM, Boyar FZ, Driscoll DJ (2004) Dynamic histone modifications mark sex chromosome inactivation and reactivation during mammalian spermatogenesis. Proc Natl Acad Sci USA 101:16583–16587
Takada Y, Isono K, Shinga J, Turner JM, Kitamura H et al (2007) Mammalian Polycomb Scmh1 mediates exclusion of Polycomb complexes from the XY body in the pachytene spermatocytes. Development 134:579–590
La Salle S, Sun F, Zhang XD, Matunis MJ, Handel MA (2008) Developmental control of sumoylation pathway proteins in mouse male germ cells. Dev Biol 321:227–237
Kim S, Namekawa SH, Niswander LM, Ward JO, Lee JT et al (2007) A mammal-specific Doublesex homolog associates with male sex chromatin and is required for male meiosis. PLoS Genet 3:e62
Chizaki R, Yao I, Katano T, Matsuda T, Ito S (2011) Restricted expression of Ovol2/MOVO in XY body of mouse spermatocytes at the pachytene stage. J Androl 33:277–286
Bao J, Wu Q, Song R, Jie Z, Zheng H et al (2011) RANBP17 is localized to the XY body of spermatocytes and interacts with SPEM1 on the manchette of elongating spermatids. Mol Cell Endocrinol 333:134–142
Tsutsumi M, Kogo H, Kowa-Sugiyama H, Inagaki H, Ohye T et al (2011) Characterization of a novel mouse gene encoding an SYCP3-like protein that relocalizes from the XY body to the nucleolus during prophase of male meiosis I. Biol Reprod 85:165–171
Costa Y, Speed RM, Gautier P, Semple CA, Maratou K et al (2006) Mouse MAELSTROM: the link between meiotic silencing of unsynapsed chromatin and microRNA pathway? Hum Mol Genet 15:2324–2334
Miura K, Imaki J (2006) Phosphorylated extracellular signal-regulated kinase 1/2 is localized to the XY body of meiotic prophase spermatocytes. Biochem Biophys Res Commun 346:1261–1266
Luijsterburg MS, Dinant C, Lans H, Stap J, Wiernasz E et al (2009) Heterochromatin protein 1 is recruited to various types of DNA damage. J Cell Biol 185:577–586
Pei H, Zhang L, Luo K, Qin Y, Chesi M et al (2011) MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature 470:124–128
Wu S, Rice JC (2011) A new regulator of the cell cycle: the PR-Set7 histone methyltransferase. Cell Cycle 10:68–72
Scharf AN, Meier K, Seitz V, Kremmer E, Brehm A et al (2009) Monomethylation of lysine 20 on histone H4 facilitates chromatin maturation. Mol Cell Biol 29:57–67
Cocquet J, Ellis PJ, Yamauchi Y, Mahadevaiah SK, Affara NA et al (2009) The multicopy gene Sly represses the sex chromosomes in the male mouse germline after meiosis. PLoS Biol 7:e1000244
Reynard LN, Turner JM (2009) Increased sex chromosome expression and epigenetic abnormalities in spermatids from male mice with Y chromosome deletions. J Cell Sci 122:4239–4248
Soboleva TA, Nekrasov M, Pahwa A, Williams R, Huttley GA et al (2011) A unique H2A histone variant occupies the transcriptional start site of active genes. Nat Struct Mol Biol 19:25–30
Tan M, Luo H, Lee S, Jin F, Yang JS et al (2011) Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146:1016–1028
Montellier E, Rousseaux S, Zhao Y, Khochbin S (2011) Histone crotonylation specifically marks the haploid male germ cell gene expression program: post-meiotic male-specific gene expression. Bioessays 34:187–193
McKee BD, Handel MA (1993) Sex chromosomes, recombination, and chromatin conformation. Chromosoma 102:71–80
Huynh KD, Lee JT (2003) Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature 426:857–862
Namekawa SH, Payer B, Huynh KD, Jaenisch R, Lee JT (2010) Two-step imprinted X inactivation: repeat versus genic silencing in the mouse. Mol Cell Biol 30:3187–3205
Okamoto I, Arnaud D, Le Baccon P, Otte AP, Disteche CM et al (2005) Evidence for de novo imprinted X-chromosome inactivation independent of meiotic inactivation in mice. Nature 438:369–373
Mahadevaiah SK, Royo H, VandeBerg JL, McCarrey JR, Mackay S et al (2009) Key features of the X inactivation process are conserved between marsupials and eutherians. Curr Biol 19:1478–1484
Ellegren H (2011) Sex-chromosome evolution: recent progress and the influence of male and female heterogamety. Nat Rev Genet 12:157–166
Acknowledgments
We thank Paul R. Andreassen and Yuya Ogawa for discussion and helpful comments regarding the manuscript. Y. I. is a research fellow of the Japan Society for the Promotion of Science. This work was supported by the Developmental Fund and Trustee Grant at Cincinnati Children’s Hospital Medical Center, the Basil O’Connor Award from the March of Dimes Foundation, and NIH Grant GM098605 to S.H.N.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ichijima, Y., Sin, HS. & Namekawa, S.H. Sex chromosome inactivation in germ cells: emerging roles of DNA damage response pathways. Cell. Mol. Life Sci. 69, 2559–2572 (2012). https://doi.org/10.1007/s00018-012-0941-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-012-0941-5