Abstract
Bacterial spores are robust and dormant life forms with formidable resistance properties, in part, attributable to the multiple layers of protein that encase the spore in a protective and flexible shield. The coat has a number of features pertinent to the emerging field of nanobiotechnology including self-assembling protomers and the capacity for engineering and delivery of foreign molecules. This review gives an account of recent progress describing the use of the spore, and specifically, the spore coat as a vehicle for heterologous antigen presentation and protective immunization (vaccination). As interest in the spore coat increases it seems likely that they will be exploited further for drug and enzyme delivery as well as a source of novel self-assembling proteins.
Similar content being viewed by others
Background
Nanostructured surfaces exhibit unique physical and chemical properties that can be exploited for many important technological applications to produce molecular structures and systems for the assembly within the nanometre size range. Nanotechnology not only can produce surfaces with novel functionality, but also new devices that are cheaper and faster than conventional ones, and which may have other advantages. There are numerous biological applications of nanotechnology, including self-assembly of supramolecular structures, slow release and delivery of enzymes and drugs, biocoatings and molecular switches actuated by chemicals, electrons or light. Many of these applications involve the development of sophisticated self-assembled surface substrates, particularly those with defined spacing. The emerging science of nanobiotechnology relies on the observation that, through evolution, nature has produced highly complex nanostructures using macromolecules, especially nucleic acids, polysaccharides and proteins. Accordingly, by understanding the principles of how these macromolecules interact to produce nanostructures, it should be possible to exploit this knowledge in the design and synthesis of new artificial structures and devices. The advantages of this "learning from nature" approach is that well defined fabrication processes already exist in bacteria.
Many organisms, particularly microorganisms, have novel and interesting structures that could be exploited, for example, the lattice-type crystalline arrays of bacterial S-layers [1, 2] and bacterial spore coats [3] both of which have protective properties. This review will examine recent studies exploiting the bacterial spore as a vaccine vehicle where the spore coat has been used for the display of heterologous antigens. In principle, the spore coat could be used not only as a delivery vehicle for a variety of different molecules but also as a source of new and novel self-assembling proteins.
Surface Display Systems
Biological applications of surface display systems are numerous and include the development of bioadsorbents and biocatalysts, the identification of new antibiotics and antigens, and the delivery of vaccines and drugs. While certainly not complete, this illustrates some of the potentially exciting and challenging areas that have recently attracted researchers' attention worldwide. Proteins able to bind metal ions or other pollutants as well as enzymes able to degrade polysaccharides when expressed in heterologous hosts can be used as bioadsorbents for heavy metal removal [4, 5], as whole-cell biocatalysts for detoxification of harmful organic contaminants from the environment [6], or for polysaccharide-degradation by the food and paper pulp industry. Peptides expressed in appropriate hosts are used to obtain combinatorial libraries and then used for i) the determination of epitope specificity of monoclonal antibodies, ii) the identification of interacting proteins and interaction sites [7] and iii) the identification of new antibiotics [8–10]. Proteins and peptides with high antigenic or pharmacological activity once expressed in appropriate hosts have been used for the development of new vaccines and drugs [11–13].
Several approaches have been undertaken to develop efficient display systems expressing heterologous polypeptides on the surface of viruses [14], microbial [7, 15], mammalian [16] and insect [17] cells. All these systems share a common theme, targeting recombinant proteins to the cell surface by constructing gene fusions using sequences from membrane-anchoring domains of surface proteins [18]. Each surface display system involves at least two components: a carrier protein, anchored on the cell surface, and a heterologous passenger protein exposed outside the cell. Several characteristics of carrier proteins can affect the efficiency of surface display and have different effects on the stability and integrity of the host cell. A successful carrier protein should meet the following requirements: i) it should have an efficient signal peptide or transporting signal, to allow the fusion protein to go through the inner membrane; ii) it should have a strong anchoring motif, to avoid detachment from the surface; and iii) it should be resistant to the proteases present in the extracellular medium or in the periplasmic space.
The location of insertion, or fusion, of the heterologous protein into the carrier protein is another important factor, since it can influence the stability, the activity and post-translational modification of the fusion protein. Therefore, fusions at the N-terminus, C-terminus or interior of the carrier protein (sandwich fusions) are, in some cases, constructed with the same passenger to obtain an efficient display. Some characteristics of the passenger protein can also affect the translocation process and final surface display. The folding structure of the passenger protein such as the formation of disulfide bridges at the periplasmic side of the outer membrane [19] or the presence of many charged or hydrophobic residues can affect translocation through the membrane [20]. The choice of the host organism is also an essential step for efficient display. A good host should be compatible with proteins to be displayed and should be easy to manipulate and cultivate without cell lysis.
Bacterial Spore Coats
Spore coats are comprised of protein, have ordered arrays of protomeric subunits, exhibit self-assembly and have protective properties [3]. As dormant metabolically inactive life forms, spores can survive indefinitely in a desiccated state, and indeed have been documented as surviving intact for millions of years [21, 22]. The spore can resist temperatures as high as 90°C as well as exposure to noxious chemicals [23]. Most (but not all) spore forming bacteria belong to two principal genera, Bacillus and Clostridium. Clostridia spore-formers, unlike Bacillus, only differentiate under anaerobic conditions making Bacillus the most amenable genus for study.
Bacillus species produce a single spore or endospore (as opposed to fungal exospores), within the bacterial cell by a process of differentiation requiring the coordinated action of hundreds of developmental genes [24, 25]. Typically mature spores are 0.8–1.2 μm in length and have either a spherical or ellipsoidal shape (see Figure 1A). The single bacterial chromosome is condensed within the centre of the spore known as the core. Layers of lipid membrane and modified peptidoglycan surround the spore core but the most important structure is the spore coat. This laminated proteinaceous shell provides the spore with resistance to organic solvents and lysozyme. In Bacillus subtilis as many as 25 different coat proteins are present in two distinct coat layers (Figure 1B), the inner and outer coat, but in other species there is evidence that the coat is less complex and may in some cases consist of only a few protein types [3, 26, 27]. The structure and assembly of the spore coat is now emerging as a model system to understand complex morphogenetic assembly processes akin to classical studies on phage T4 assembly. The outer, electron-dense, layer of the B. subtilis coat is comprised of 5 principal polypeptides, CotA (65 kDa), CotB (59 kDa), CotG (24 kDa), CotC (11 kDa) and CotF (8 kDa). CotA is a multi-copper oxidase [28, 29] and can accumulate in multimeric forms (observed microscopically) within the sporulating cells in some coat-defective mutants [3]. Presumably oligomerisation and self-assembly of CotA precedes deposition onto the spore coat surface. The CotG and CotB proteins have also been shown to interact covalently [27] and in addition CotG [30] and also CotC [31, 32] have extremely unusual amino acid sequences containing multiple repeats (>13) of 12–13 amino acids rich in lysines and tyrosines. Furthermore, many of the spore coat proteins have unusual profiles i.e. multimeric forms and aberrant molecular masses, when examined by SDS-PAGE. Recently, it has been shown that the spore coat is actually flexible and can expand and contract and this feature is critical for spore formation when the spore dehydrates and similarly for germination when the spore rehydrates [33, 34]. This aspect of a self-assembled structure is particulary interesting and might offer a number of future applications in drug delivery, nanofabrication and surface coatings.
Bacterial endospores. Panel A shows endospores from B. subtilis one of which is still retained within the rod shaped 'mother cell'. In B. subtilis, spores are approximately 1.2 μm in length and are ellipsoidal. Released spores have a clear protective shell known as the spore coat and is comprised of as many as 25 different protomeric components assembled into discrete layers. Panel B shows a typical SDS-PAGE (12.5%) fractionation of solubilised spore coat proteins revealing predominant species.
Engineering the Bacterial Spore Coat
A strategy to engineer Bacillus subtilis spores to display heterologous antigens on the spore surface has been recently reported [35] and is illustrated in Figure 2. A spore-based display system provides several advantages with respect to systems based on the use of bacterial cells, these include the robustness of the bacterial spore allowing storage in the desiccated form, ease of production, safety and a technological platform supported by extensive tools for genetic manipulation.
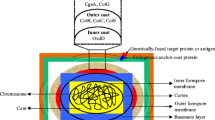
Spore-surface display using spore coat proteins. The B. subtilis spore is composed of an internal core (yellow) surrounded by a peptidoglycan-like cortex (in red) and a proteinaceous coat sub-divided into an inner (gree) and an outer (black) part. The fusion protein, composed by a carrier (blue) and a passenger (purple) part is exposed on the spore surface.
In contrast to the wealth of information available on how gene expression controls differentiation of a growing cell into a dormant spore little is known about the mechanisms of protein incorporation into the coat, the nature of structural components forming the most external part of the coat and whether there are anchoring motifs. Initial attempts to expose heterologous proteins on the spore surface have been focused on two coat components, CotB and CotC. In the case of CotB this spore coat protein is known to be surface located [35] while for CotC, relative to other coat proteins, this species has a high relative abundance [36]. The observation that both of these coat components was dispensable for the formation of an apparently normal spore as well as for its germination, was an additional positive feature in choosing CotB and CotC as potential carrier proteins.
Two antigens were initially selected as model proteins to display on the spore surface: i) the non-toxic 459 amino acid C-terminal fragment of the tetanus toxin (TTFC), a well characterized and highly immunogenic 51.8 kDa peptide [37], encoded by the tetC gene of Clostridium tetani; and ii) the 103 amino acid B subunit of the heat-labile toxin of enterotoxigenic strains of Escherichia coli (LTB), a 12 kDa peptide, encoded by the eltB gene [38].
CotB as a carrier protein
Like other coat components, CotB has been associated to the outer coat layer on the basis of genetic evidence [39] and only recently an immunocytofluorimetric analysis performed on intact spores has shown that CotB is accessible to CotB-specific antibodies and therefore that it is most probably exposed on the spore surface [35].
The CotB structural gene, cotB, is under the dual transcriptional control of σK and the DNA-binding protein GerE. As a consequence, cotB is transcribed only in the mother cell compartment of the sporulating cell [27]. Once synthesised in the mother cell cytoplasm, CotB is assembled around the forming spore in a manner somehow dependent on CotE, CotG and CotH. Therefore, CotB and the heterologous protein eventually fused to it, do not undergo a cell wall translocation step, typical of display systems in other bacteria.
CotB has a strongly hydrophilic C-terminal half formed by three 27 amino acid-repeats rich in serine, lysine and glutamine residues. Serine residues account for over 50% of the CotB C-terminal half. The lysine residues in the CotB repeats have been suggested to represent sites of intra- or inter-molecular cross-linking, by analogy to the connective tissue proteins collagen and elastin [27, 40]. The CotB protein has a deduced molecular mass of 46 kDa, but migrates on SDS-PAGE as a 59 kDa polypeptide. Recently the discrepancy between measured and deduced molecular weight has been explained by showing that CotB is initially synthesized as a 46 kDa species, and converted into a 59 kDa homodimer [41], that retains both the N- an C-terminal ends predicted from the cotB nucleotide sequence.
The strategy to obtain recombinant B. subtilis spores expressing CotB-TTFC or CotB-LTB on their surface was based on (i) use of the cotB gene and its promoter for the construction of translational fusions and (ii) chromosomal integration of the cotB-tetC and cotB-eltB gene fusions into the coding sequence of the non-essential gene amyE (Figure 3A) [42]. Placing the fusion proteins under cotB transcriptional and translational signals ensured correct timing of expression during sporulation, while its chromosomal integration guaranteed the genetic stability of the construct. Due to the lack of information on CotB coat assembly and on the requirements for anchoring motifs, initial attempts were performed by positioning the passenger protein at the C-terminal, the N-terminal or in the middle of CotB (Fig. 3B).
Spore-surface display system in B. subtilis . (A) Schematic view of the integration of gene fusions on the chromosome. Red bars represent the gene fusion, grey and green bars represent respectively the non-essential amyE gene of B. subtilis used as integrational site and the chloramphenicol acetyl transferase (cat) gene used as a selectable marker. (B) Schematic representation of fusion proteins using either full length CotB (380 amino acids) or CotBΔ105 (275 amino acids) or CotC (66 amino acids) as carrier proteins (all in blue). The three 27-amino-acids repeats of full lenght CotB and the part of them remaining in CotBΔ105 are in black. Purple bars represent the heterologous part of the fusions (TTFC or LTB).
When TTFC and LTB were fused to the C-terminal end of CotB, the chimeric proteins failed to correctly assemble on the spore surface (Isticato and Ricca, unpublished). Such initial failures were attributed to a potential instability of the constructs, either at the DNA level (repetitive DNA sequences) or at the protein level. In order to bypass such problems TTFC and LTB were fused to the C-terminal end of a CotB form deleted of the three 27 amino acid-repeats, CotBΔ105-TTFC (Fig. 3A). In contrast to the full length version, the CotBΔ105-TTFC chimeric protein was correctly assembled and exposed on the spore surface [35]. A quantitative dot blot showed that each recombinant spore exposed an amount of CotBΔ105-TTFC fusion protein equal to 0.00022 pg making it possible to conclude that 1.5 × 103 chimeric molecules are present on the surface of each recombinant spore [35].
Unlike CotBΔ105-TTFC, CotBΔ105-LTB was not properly assembled. The strain expressing this chimera showed reduced sporulation and germination efficiencies and its spores were not resistant to lysozyme. These observations, together with the SDS-PAGE analysis of the released coat proteins, suggested that the presence of CotBΔ105-LTB strongly altered the spore coat layer. An in-silico analysis showed some homology between the chimeric product (in the fusion region) and LytF, a cell wall-associated endopeptidase produced by B. subtilis during vegetative growth, thus raising the possibility that the chimeric product could interfere with proper coat formation by degrading some coat components (Mauriello and Ricca, data not shown).
In addition to the C-terminal end fusion described above the model passenger protein TTFC has been fused also at the N-terminal and in the middle of CotB (Fig. 3B). In both cases the CotBΔ105 form of CotB was used to avoid the problems experienced with the C-terminal fusion (see above). Both the N-terminal and the sandwich fusions originated chimeric products that were properly assembled in the coat structure from both the qualitative and the quantitative point of view [35]. At least in the case of CotB, it was then possible to conclude that where the passenger protein is exposed it does not affect display on the spore surface.
CotC as a carrier protein
CotC is a 12 kDa, alkali-soluble component of the B. subtilis spore coat, previously identified by reverse genetics [32] and then associated to the outer coat layer based on genetic evidence [39]. CotC was initially considered as a carrier candidate because of its relative abundance in the coat (Figure 1B). Together with CotG and CotD, CotC represents about 50% of the total solubilized coat proteins. Such relatively high amounts could allow the assembly of a significant number of CotC-based chimeras on the coat, thus ensuring an efficient heterologous display. Expression of the cotC gene is under the control of the mother cell specific σ-factor σK and of transcriptional regulators GerE and SpoIIID. As in the case of CotB, CotC is also transcribed in the mother cell and its assembly on the coat does not require membrane translocation. The primary product of the cotC gene is a 66 amino acid polypeptide extremely rich in tyrosine (30.3%) and lysine (28.8%) residues [32]. However, it was recently shown that CotC is assembled into at least four distinct protein forms, ranging in size between 12 and 30 kDa [31]. Two of these, having molecular masses of 12 and 21 kDa and corresponding most likely to a monomeric and homodimeric form of CotC respectively, are assembled on the forming spore right after their synthesis eight hours after the onset of sporulation. The other two forms, 12.5 and 30 kDa, are probably the products of post-translational modifications of the two other forms, occurring directly on the coat surface during spore maturation [31].
In the case of CotC only C-terminal fusions have been so far constructed (Figure 3B). Both CotC-TTFC and CotC-LTB gene fusions were obtained by cloning tetC or eltB in frame with the last cotC codon under the transcriptional and translational control of the cotC promoter region. The gene fusion was then integrated into the B. subtilis chromosome at the amyE locus by double cross-over recombination [36] (Figure 3A). Both these two chimeric proteins were assembled on the coat of recombinant spores without major effect on the spore structure and/or function, since they appeared identical to wild type spores in terms of efficiency of sporulation and germination and resistance properties. Western blot, cytofluorimetric analysis and, for CotC-TTFC, immunofluorescence microscopy (Figure 4) showed that both CotC-based chimeras were displayed on the surface of the recombinant spores. A quantitative determination of recombinant proteins exposed on B. subtilis spores revealed that ca. 9.7 × 102 and 2.7 × 103 molecules of CotC-TTFC and CotC-LTB, respectively, were extracted from each spore.
Detection of presence of CotC-LTB and CotC-TTFC by immunofluorescence microscopy. Sporulation of B. subtilis strains was induced by the resuspension method, and samples were taken 6 h after the onset of sporulation and analysed by immunofluorescence microscopy as described previously [46]. Samples were labelled with mouse anti-LTB antibody followed by anti-mouse IgG-TRITC conjugate (red fluorescein, Panels A & B), or rabbit anti-TTFC antibody followed by anti-rabbit IgG-FITC conjugate (green fluorescein, Panels C & D). Panels A & C, wild type spores; Panel B, isogenic spores expressing CotC-LTB); Panel D, isogenic spores expressing CotC-TTFC.
Although CotC appears more abundant than CotB within the coat, comparable amounts of heterologous proteins are exposed by the CotC-based and the CotBΔ105-based systems. This result was somewhat unexpected, since CotC appears to be much more abundant than CotB in the coat. A possible explanation comes from the recent finding that the C-terminal end of CotC is not only essential for interaction with other CotC molecules but also to other coat components (Isticato and Ricca, manuscript in preparation) and therefore shows that the use of CotC as a carrier still needs to be optimised.
Stability of Spore-Displayed Proteins
One of the main reasons to propose the use of the bacterial spore as a favourable display system is its well-documented stability. Spores can be simply stored at room temperature for a long time without reduction of their resistance and stability properties. This would be an extremely useful property for a variety of biotechnological applications. As an example, if the passenger protein is an antigen, the recombinant spore could become an ideal heat-stable oral vaccine for use in developing countries, where heat-stability is of most concern due to poor distribution and storage.
However, while spore stability is well documented [23], stability of heterologous proteins exposed on the spore surface has only recently been investigated. Spores expressing CotBΔ105-TTFC (see above) and parental spores were stored at -80°C, -20°C, +4°C and at room temperature and assayed at different storage times up to 12 weeks. In all cases the amount of heterologous protein present on the surface of recombinant spores appeared identical between freshly prepared spores and those stored for up to 12 weeks (Fig. 5). These results, indicating that heterologous proteins can be stably exposed on the surface of recombinant spores, confirm the spore-based system as a very promising display approach that could overcome some drawbacks of other systems and that could find applications in a variety of diverse biotechnological fields.
Dot blot analysis of proteins extracted from wild-type spores and from spores exposing the CotBΔ 105 -TTFC chimera. Freshly purified spores expressing the CotBΔ105-TTFC chimera (t0) or after 4, 8 or 12 weeks (t4, t8 and t12) of storage at room temperature were examined by dot blotting of extracted spore coat proteins. Concentrations of purified TTFC (in nanograms) and of coat proteins (in micrograms) are indicated. Proteins were loaded and reacted with monoclonal anti-TTFC antibodies (Boeringher), then with phosphatase-conjugated secondary antibodies. Signals of similar intensity were obtained with freshly purified spores and spores stored at room temperature for up to 12 weeks.
Proof of Principle for Oral Vaccination Using Tetanus as a Model Disease
Spores expressing CotBΔ105-TTFC have been used to immunise mice by the oral route [43]. Serum IgG and faecal sIgA showed clear seroconversion to TTFC. The dosing schedule used three sets of three doses (1.67 × 1010) over 5 weeks and was based on regimes optimised for oral immunisations [44, 45]. The titres of TTFC-specific IgG after 33 days (>103) suggested that these were at protective levels and mice challenged with tetanus toxin corresponding to 10 LD50 were fully protected. Out of eight mice challenged with a 20 LD50 dose seven survived suggesting that this was the threshold for protection. A similar study was made using nasal immunisation with CotB-TTFC spores but with a lower dose and three immunisations [43]. Here, TTFC-specific IgG responses were lower but still showed seroconversion. These studies show that engineered spores expressing a heterologous antigen can be used for protective immunisation. Moreover, although mucosal responses are not important for protection against Clostridium tetani (a systemic pathogen) they are obviously important for mucosal pathogens. Further studies will be needed to optimise dosing regimes (less doses and less spores) but these seminal studies have opened the way for development against specific mucosal pathogens. Although these studies are encouraging and demonstrate humoral responses there is no clear evidence yet that indicates cellular responses. However, spores have been shown to disseminate to the GALT and are found in the Peyer's Patches (PP) and mesenteric lymph nodes [46]. The small size of the spore (1 μm) would allow it to be taken up by M cells and transported to the PP where it could interact with antigen presenting cells. Initial studies have shown that spores can germinate and persist for a short period of time within intestinal macrophages as well as elicit Th 1 cytokines in vivo such as IFN-γ [47].
Conclusions
The protective coat of the bacterial spore offers two areas of interest for long-term exploitation pertinent to nanobiotechnology. First, as a substrate for delivery of biomolecules and second as a source for understanding self-assembling molecules. As a delivery vehicle the spore coat has been shown to provide a suitable surface for display of heterologous antigens using the CotB and CotC proteins. As vaccine vehicles spores have a number of advantages over other second generation systems under development. However, the same carried systems (CotB and CotC) could also be used for delivery of proteins important for industry (e.g. xylanases) or for drug delivery. It is probable that other spore coat proteins can be used for surface expression and delivery and indeed spore-forming species other than B. subtilis could be developed. The spore coat carries scores of different protomers of which some self-assemble although little is currently known about how this occurs. Accordingly, the assembly processes that form the spore coat as well as the nature of individual proteins could provide a rich source of hitherto unknown self-assembling molecules.
References
Sidhu MS, Olsen I: S-layers of Bacillus species. Microbiol. 1997, 143: 1039-1052.
Sleytr UB, Messner P: Crystalline surface layers on bacteria. Ann Rev Microbiol. 1983, 37: 311-339. 10.1146/annurev.mi.37.100183.001523.
Driks A: Bacillus subtilis spore coat. Microbiol Mol Biol Rev. 1999, 63: 1-20.
Kotrba P, Doleckova L, de Lorenzo V, Ruml T: Enhanced bioaccumulation of heavy metal ions by bacterial cells due to surface display of short metal binding peptides. Appl Environ Microbiol. 1999, 65 (3): 1092-1098.
Samuelson P, Wernerus H, Svedberg M, Stahl S: Staphylococcal surface display of metal-binding polyhistidyl peptides. Appl Environ Microbiol. 2000, 66 (3): 1243-1248. 10.1128/AEM.66.3.1243-1248.2000.
Dhillon JK, Drew PD, Porter AJ: Bacterial surface display of an anti-pollutant antibody fragment. Lett Appl Microbiol. 1999, 28 (5): 350-354. 10.1046/j.1365-2672.1999.00552.x.
Georgiou G, Stathopoulos C, Daugherty PS, Nayak AR, Iverson BL, Curtiss R: Display of heterologous proteins on the surface of microorganisms: from the screening of combinatorial libraries to live recombinant vaccines. Nat Biotechnol. 1997, 15 (1): 29-34.
Murase K, Morrison KL, Tam PY, Stafford RL, Jurnak F, Weiss GA: EF-Tu binding peptides identified, dissected, and affinity optimized by phage display. Chem Biol. 2003, 10 (2): 161-168. 10.1016/S1074-5521(03)00025-5.
Kim WC, Rhee HI, Park BK, Suk KH, Cha SH: Isolation of peptide ligands that inhibit glutamate racemase activity from a random phage display library. J Biomol Screen. 2000, 5 (6): 435-440.
Huang W, Zhang Z, Palzkill T: Design of potent beta-lactamase inhibitors by phage display of beta-lactamase inhibitory protein. J Biol Chem. 2000, 275 (20): 14964-14968. 10.1074/jbc.M001285200.
Liljeqvist S, Samuelson P, Hansson M, Nguyen TN, Binz H, Stahl S: Surface display of the cholera toxin B subunit on Staphylococcus xylosus and Staphylococcus carnosus. Appl Environ Microbiol. 1997, 63 (7): 2481-2488.
Lee JS, Shin KS, Pan JG, Kim CJ: Surface-displayed viral antigens on Salmonella carrier vaccine. Nat Biotechnol. 2000, 18 (6): 645-648. 10.1038/76494.
Chen H, Schifferli DM: Construction, characterization, and immunogenicity of an attenuated Salmonella enterica serovar typhimurium pgtE vaccine expressing fimbriae with integrated viral epitopes from the spiC promoter. Infect Immun. 2003, 71 (8): 4664-4673. 10.1128/IAI.71.8.4664-4673.2003.
Rahman MM, Gopinathan KP: Bombyx mori nucleopolyhedrovirus-based surface display system for recombinant proteins. J Gen Virol. 2003, 84 (Pt 8): 2023-2031. 10.1099/vir.0.19225-0.
Stahl S, Uhlen M: Bacterial surface display: trends and progress. Trends Biotechnol. 1997, 15 (5): 185-192. 10.1016/S0167-7799(97)01034-2.
Whitehorn EA, Tate E, Yanofsky SD, Kochersperger L, Davis A, Mortensen RB, Yonkovich S, Bell K, Dower WJ, Barrett RW: A generic method for expression and use of "tagged" soluble versions of cell surface receptors. Biotechnology (NY). 1995, 13 (11): 1215-1219.
Ernst W, Grabherr R, Wegner D, Borth N, Grassauer A, Katinger H: Baculovirus surface display: construction and screening of a eukaryotic epitope library. Nucleic Acids Res. 1998, 26 (7): 1718-1723. 10.1093/nar/26.7.1718.
Lee SY, Choi JH, Xu Z: Microbial cell-surface display. Trends Biotechnol. 2003, 21 (1): 45-52. 10.1016/S0167-7799(02)00006-9.
Maurer J, Jose J, Meyer TF: Autodisplay: one-component system for efficient surface display and release of soluble recombinant proteins from Escherichia coli. J Bacteriol. 1997, 179 (3): 794-804.
Nguyen TN, Gourdon MH, Hansson M, Robert A, Samuelson P, Libon C, Andreoni C, Nygren PA, Binz H, Uhlen M: Hydrophobicity engineering to facilitate surface display of heterologous gene products on Staphylococcus xylosus. J Biotechnol. 1995, 42 (3): 207-219. 10.1016/0168-1656(95)00081-Z.
Nicholson WL: Roles of Bacillus endospores in the environment. Cell Mol Life Sci. 2002, 59: 410-416. 10.1007/s00018-002-8433-7.
Cano RJ: Borucki M Revival and identification of bacterial spores in 25- to 40-million-year-old Dominican amber. Science. 1995, 268: 1060-1064.
Nicholson WJ, Munakata N, Horneck G, Melosh HJ, Setlow P: Resistance of Bacillus endospores to extreme terrestial and extraterrestrial environments. Microbiol Mol Biol Rev. 2000, 64: 548-572. 10.1128/MMBR.64.3.548-572.2000.
Errington J: Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol Rev. 1993, 57 (1): 1-33.
Errington J: Regulation of endospore formation in Bacillus subtilis. Nature Reviews Microbiology. 2003, 1: 117-126. 10.1038/nrmicro750.
Aronson AI, Fitz-James P: Structure and morphogenesis of the bacterial spore coat. Bacteriol Rev. 1976, 40 (2): 360-402.
Henriques AO, Moran CP: Structure and assembly of the bacterial endospore coat. Methods, a Companion to Methods in Enzymology. 2000, 20: 95-110. 10.1006/meth.1999.0909.
Martins LO, Soares CM, Pereira MM, Teixeira M, Costa T, Jones GH, Henriques AO: Molecular and biochemical characterization of a highly stable bacterial laccase that occurs as a structural component of the Bacillus subtilis endospore coat. J Biol Chem. 2002, 277 (21): 18849-18859. 10.1074/jbc.M200827200.
Hullo MF, Moszer I, Danchin A, Martin-Verstraete I: CotA of Bacillus subtilis is a copper-dependent laccase. J Bacteriol. 2001, 183 (18): 5426-5430. 10.1128/JB.183.18.5426-5430.2001.
Sacco M, Ricca E, Losick R, Cutting S: An additional GerE-controlled gene encoding an abundant spore coat protein from Bacillus subtilis. J Bacteriol. 1995, 177: 372-377.
Isticato R, Esposito G, Zilhao R, Nolasco S, Cangiano G, De Felice M, Henriques AO, Ricca E: Assembly of multiple CotC forms into the Bacillus subtilis spore coat. J Bacteriol. 2004,
Donovan W, Zheng L, Sandman K, Losick R: Genes encoding spore coat polypeptides from Bacillus subtilis. J Mol Biol. 1987, 196: 1-10.
Driks A: The dynamic spore. Proc Natl Acad Sci U S A. 2003, 100 (6): 3007-3009. 10.1073/pnas.0730807100.
Chada VGR, Sanstad EA, Wang R, Driks A: Morphogenesis of Bacillus spore surfaces. J Bacteriol. 2003, 185: 6255-6261. 10.1128/JB.185.21.6255-6261.2003.
Isticato R, Cangiano G, Tran HT, Ciabattini A, Medaglini D, Oggioni MR, De Felice M, Pozzi G, Ricca E: Surface display of recombinant proteins on Bacillus subtilis spores. J Bacteriol. 2001, 183: 6294-6301. 10.1128/JB.183.21.6294-6301.2001.
Mauriello EMF, Duc LH, Isticato R, Cangiano G, Hong HA, De Felice M, Ricca E, Cutting SM: Display of Heterologous Antigens on the Bacillus subtilis Spore Coat Using CotC as a Fusion Partner. Vaccine. 2003,
Helting TB, Zwisler O: Structure of tetanus toxin. I. Breakdown of the toxin molecule and discrimination between polypeptide fragments. J Biol Chem. 1977, 252: 194-198.
Douce G, Turcotte C, Cropley I, Roberts M, Pizza M, Domenghini M, Rappuoli R, Dougan G: Mutants of Escherichia coli heat-labile toxin lacking ADP-ribosyltransferase activity act as nontoxic, mucosal adjuvants. Proc Natl Acad Sci U S A. 1995, 92: 1644-1648.
Zheng L, Donovan WP, Fitz-James PC, Losick R: Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Dev. 1988, 2: 1047-1054.
Kobayashi K, Suzuki SI, Izawa Y, Miwa K, Yamanaka S: Transglutaminase in sporulating cells of Bacillus subtilis. J Gen Appl Microbiol. 1998, 44 (1): 85-91.
Zilhao R, Isticato R, Ozin AJ, Serrano M, Moran CP, Ricca E, Henriques AO: Interactions among CotB, CotG, and CotH during assembly of the Bacillus subtilis spore coat. J Bacteriol. 2004,
Cutting SM, Vander-Horn PB: Genetic Analysis. In Molecular Biological Methods for Bacillus. Edited by: Harwood CR, Cutting SM. 1990, John Wiley & Sons Ltd., Chichester, England, 27-74.
Duc LH, Hong HA, Fairweather N, Ricca E, Cutting SM: Bacterial spores as vaccine vehicles. Infect Immun. 2003, 71: 2810-2818. 10.1128/IAI.71.5.2810-2818.2003.
Challacombe SJ: Salivary antibodies and systemic tolerance in mice after oral immunisation with bacterial antigens. Ann N Y Acad Sci. 1983, 409: 177-192.
Robinson K, Chamberlain LM, Schofield KM, Wells JM, Le Page RWF: Oral vaccination of mice against tetanus with recombinant Lactococcus lactis. Nat Biotechnol. 1997, 15: 653-657.
LH D, Hong HA, Fairweather N, Ricca E, Cutting SM: Bacterial spores as vaccine vehicles. Infect Immun. 2003, 71 (5): 2810-2818. 10.1128/IAI.71.5.2810-2818.2003.
Duc LH, Hong HA, Uyen NQ, Cutting SM: Immunogenicity and intracellular fate of B. subtilis spores. Vaccine. 2004,
Acknowledgements
This work was supported by grants from the Wellcome Trust (SMC) and the European Union to SMC and ER.
Author information
Authors and Affiliations
Corresponding author
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Ricca, E., Cutting, S.M. Emerging Applications of Bacterial Spores in Nanobiotechnology. J Nanobiotechnol 1, 6 (2003). https://doi.org/10.1186/1477-3155-1-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1477-3155-1-6