Abstract
Wild sorghums are extremely diverse phenotypically, genetically and geographically. However, there is an apparent lack of knowledge on the genetic structure and diversity of wild sorghum populations within and between various eco-geographical regions. This is a major obstacle to both their effective conservation and potential use in breeding programs. The objective of this study was to assess the genetic diversity and structure of wild sorghum populations across a range of eco-geographical conditions in Kenya. Sixty-two wild sorghum populations collected from the 4 main sorghum growing regions in Kenya were genotyped using 18 simple sequence repeat markers. The study showed that wild sorghum is highly variable with the Coast region displaying the highest diversity. Analysis of molecular variance showed a significant variance component within and among wild sorghum populations within regions. The genetic structure of wild sorghum populations indicated that gene flow is not restricted to populations within the same geographic region. A weak regional differentiation was found among populations, reflecting human intervention in shaping wild sorghum genetic structure through seed-mediated gene flow. The sympatric occurrence of wild and cultivated sorghums coupled with extensive seed-mediated gene flow, suggests a potential crop-to-wild gene flow and vice versa across the regions. Wild sorghum displayed a mixed mating system. The wide range of estimated outcrossing rates indicate that some environmental conditions may exist where self-fertilisation is favoured while others cross-pollination is more advantageous.





Similar content being viewed by others
References
Anas, Yoshida T (2004) Sorghum diversity evaluated by simple sequence repeat (SSR) markers and phenotypic performance. Plant Prod Sci 7:301–308
Anttila CK, Daehler CC, Rank NE, Strong DR (1998) Greater male fitness of a rare invader (Spartina alterniflora, Poaceae) threatens a common native (Spartina foliosa) with hybridization. Am J Bot 85:1597–1601
Ayana A, Bekele E (1998) Geographical patterns of morphological variation in sorghum (Sorghum bicolor (L.) Moench) germplasm from Ethiopia and Eritrea: qualitative characters. Hereditas 129:195–205
Ayana A, Bryngelsson T, Bekele E (2000) Genetic variation of Ethiopian and Eritrean Sorghum (Sorghum bicolor (L.) Moench) germplasm assessed by random amplified polymorphic DNA (RAPD). Genet Resour Crop Evol 47:471–482
Barnaud A, Deu M, Garine E (2007) Local genetic diversity of sorghum in a village in northern Cameroon: structure and dynamics of landraces. Theor Appl Genet 114:237–248
Barnaud A, Triguero G, McKey D, Joly HI (2008) High outcrossing rates in fields with mixed sorghum landraces: how are landraces maintained? Heredity 101:445–452
Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (2004) GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000, Université de Montpellier II, Montpellier, France
Bohonak AJ (2002) IBD (isolation by distance): a program for analyses of isolation by distance. J Hered 93:153–154
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Brown SM, Hopkins MS, Mitchell SE, Senior ML, Wang TY, Duncan RR, Gonzales-Candelas F, Kresovich S (1996) Multiple methods for identification of polymorphic simple sequence repeats (SSRs) in sorghum (Sorghum bicolor (L.) Moench). Theor Appl Genet 93:190–198
Casa AM, Mitchell SE, Hamblin MT, Sun H, Bowers JE, Paterson AH, Aquadro CF, Kresovich S (2005) Diversity and selection in sorghum: simultaneous analyses using simple sequence repeats. Theor Appl Genet 111:23–30
Conner AJ, Glare TR, Nap JP (2003) The release of genetically modified crops into the environment: Part II. Overview of ecological risk assessment. Plant J 33:19–46
De Wet JMJ (1978) Systematics and evolution of Sorghum Sect. Sorghum (Gramineae). Am J Bot 65:477–484
De Wet JMJ, Harlan JR (1971) The origin and domestication of Sorghum bicolor. Econ Bot 25:128–135
Dillon SL, Shapter FM, Henry RJ, Cordeiro G, Izquierdo L, Lee LS (2007) Domestication to crop improvement: genetic resources for Sorghum and Saccharum (Andropogoneae). Ann Bot 100:975–989
Djè Y, Heuertz M, Lefèbvre C, Vekemans X (2000) Assessment of genetic diversity within and among germplasm accessions in cultivated sorghum using microsatellite markers. Theor Appl Genet 100:918–925
Dje´ Y, Heuertz M, Ater M, Lefebvre C, Vekemans X (2004) In situ estimation of outcrossing rate in sorghum landraces using microsatellite markers. Euphytica 138:205–212
Doggett H (1988) Sorghum, 2nd edn. Longman, London
Duvall MR, Doebley JF (1990) Restriction site variation in the chloroplast genome of Sorghum (Poaceae). Syst Bot 15:472–480
Ellstrand NC, Schierenbeck KA (2000) Hybridization as a stimulus for the evolution of invasiveness in plants? Proc Natl Acad Sci USA 97:7043–7050
Epperson BK (1993) Recent advances in correlation analysis of spatial patterns of genetic variation. Evol Biol 27:95–155
Epperson BK (2004) Multilocus estimation of genetic structure within populations. Theor Popul Biol 65:227–237
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Laval G, Schneider S (2006) An integrated software package for population genetics data analysis. Arlequin v3.1. http://cmpg.unibe.ch/software
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Folkertsma RT, Frederick H, Rattunde W (2005) The pattern of genetic diversity of Guinea-race Sorghum bicolor (L.) Moench landraces as revealed with SSR markers. Theor Appl Genet 111:399–409
Gao H, Williamson S, Bustamante CD (2007) An MCMC approach for joint inference of population structure and inbreeding rates from multi-locus genotype data. Genetics (online)
Goudet J (2002) FSTAT, a program to estimate and test gene diversity and fixation indices. (version 2 9 3 2)
Hails RS, Morley K (2005) Genes invading new populations: a risk assessment perspective. Trends Ecol Evol 20:245–252
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol 2:618–620
Harlan JR, de Wet JMJ (1972) A simplified classification of cultivated sorghum. Crop Sci 12:172–176
Hokanson KE, Ellstrand NC, Ouedragado JT, Olwenya PA, Schaals B, Reybould AF (2010) Biofortified sorghum in Africa: using problem formulation to inform risk assessment. Nat Biotech 28:900–903
IBPGR/ICRISAT (1993) Descriptors for sorghum (Sorghum bicolor (L.) Moench). International Board of Plant Genetic Resources, Rome, Italy. International Crop Research Institute for Semi-Arid Tropics, Patancheru
Kalinowski S (2005) HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol 5:187–189
Kong L, Dong J, Hart GE (2000) Characteristics, linkage-map positions and allelic differentiation of Sorghum bicolor (L.) Moench DNA simple-sequence repeats (SSRs). Theor Appl Genet 101:438–448
Levin DA, Kerster HW (1974) Gene flow in seeds plants. Evol Biol 7:139–220
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics Research Center, North Carolina State University, Raleigh
Loiselle BA, Sork VL, Nason J, Graham C (1995) Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am J Bot 82:1420–1425
Lowe A, Harris S, Ashton P (2004) Ecological genetics: design analysis and application. Blackwell Publishing, USA, pp 70–72
Mace ES, Buhariwalla HK, Crouch JH (2003) A high throughput DNA extraction protocol for molecular breeding programs. Plant Mol Biol Report 21:459a–459h
Manly BFJ (1994) Multivariate statistical methods: a primer. Chapman and Hall, New York
Mann JA, Kimber CT, Miller FR (1983) The origin and early cultivation of sorghums in Africa. Texas Agricultural Experiment Station, Bulletin 1454
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Manzelli M, Pileri L, Lacerenza N, Benedettelli S, Vecchio V (2007) Genetic diversity assessment in Somali sorghum (Sorghum bicolor (L.) Moench) accessions using microsatellite markers. Biodivers Conserv 16:1715–1730
Menz MA, Klein RR, Unruh NC, Rooney WL, Klein PE, Mullet JE (2004) Genetic diversity of public inbreds of sorghum determined by mapped AFLP and SSR markers. Crop Sci 44:1236–1244
Miller MP (2000) Tools for Population Genetic Analyses version 1.3. A Windows program for the analysis of allozyme and molecular population genetic data. Department of Biological Sciences, Northern Arizona University, Flagstaff, AZ
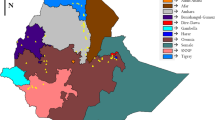
Muraya MM, Hartwig HH, Mutegi E, Kanyenji BM, Sagnard F, de Villiers S, Kiambi D, Heiko K, Parzies HK (2010) Geographical patterns of phenotypic diversity and structure of Kenyan wild sorghum populations (Sorghum spp.) as an aid to germplasm collection and conservation strategy. Plant Genet Resour 8:217–224
Muraya MM, Mutegi E, Geiger HH, de Villiers S M, Sagnard F, Kanyenji BM, Kiambi D, Parzies HK (2011) Wild sorghum from different eco-geographic regions of Kenya display a mixed mating system. Theor Appl Genet 122:1631–1639
Muteg E, Sagnard F, Muraya M, Kanyenji B, Rono B, Mwongera C, Marangu C, Kamau J, Parzies H, de Villiers S, Semagn K, Traoré PS, Labuschagne M (2010) Ecogeographical distribution of wild, weedy and cultivated Sorghum bicolor in Kenya: implications for conservation and crop-to-wild gene flow. Genet Resour Crop Evol 57:243–253
Nei M (1987) Molecular evolutionary genetics. Columbia University press, New York
Pilson D, Prendeville HR (2004) Ecological effects of transgenic crops and the escape of transgenes into wild populations. Ann Rev Ecol Evol Syst 35:149–174
Price HJ, Dillon SL, Hodnett G, Rooney WL, Ross L, Johnston JS (2005) Genome Evolution in the Genus Sorghum (Poaceae). Ann Bot 95:219–227
Pritchard JK, Stephens M, Donnely P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pritchard JK, Wen X, Falush D (2007) Documentation for structure software: Version 2.2. http://www.biovip.org/UploadFiles/2009-12/1219737299.pdf
Rabbi IY, Parzies HK, Kiambi D, Haussmann BIG, Folkertsma R, Geiger HH (2010) Experimental studies on pollen-mediated gene flow in Sorghum bicolor (L.) Moench using malesterile bait plants. Plant Breed doi:10.1111/j.1439-0523.2010.01775.x
Rich PJ, Grenier C, Ejeta G (2004) Striga resistance in the wild relatives of sorghum. Crop Sci 44:2221–2229
Rieseberg LH (1991) Homoploid reticulate evolution in Helianthus: evidence from ribosomal genes. Am J Bot 78:1218–1237
Rieseberg LH (1997) Hybrid origins of plant species. Annu Rev Ecol Syst 28:359–389
Rieseberg LH, Raymond O, Rosenthal DM, Lai Z, Livingstone K, Nakazato T, Durphy JL, Schwarzbach AE, Donovan LA, Lexer C (2003) Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301:1211–1216
Rousset F (1997) Genetic differentiation and estimation of gene flow from F statistics under isolation by distance. Genetics 145:1219–1228
Schloss SJ, Mitchell SE, White GM, Kukatla R, Bowers JE, Paterson AH, Kresovich S (2002) Characterization of RFLP probe sequences for gene discovery and SSR development in Sorghum bicolor (L.) Moench. Theor Appl Genet 105:912–920
Slatkin M (1993) Isolation by distance in equilibrium and non-equilibrium populations. Evolution 47:264–279
Snowden JD (1936) The cultivated races of Sorghum. Adlard and Son, London
Sokal RR, Oden NL (1978) Spatial autocorrelation in biology 2 Some biological implications and four applications of evolutionary and ecological interest. Biol J Linn Soc 180(10):249
Soltis DE, Soltis PS (1999) Polyploidy: recurrent formation and genome evolution. Trends Ecol Evol 14:348–352
Stewart CN, Halfhill MD, Warwick SI (2003) Genetic modification: transgene introgression from genetically modified crops to their wild relatives. Nat Rev Genet 4:806–817
Tamura K, Dudley J, Nei M, Kumar S (2006) Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Version 4.0 (Beta release) Center for Evolutionary Functional Genomics Bio design Institute, Arizona State University
Taramino G, Tarchini R, Ferrario S, Lee M, Pe` ME (1997) Characterisation and mapping of simple sequence repeats (SSR) in Sorghum bicolor. Theor Appl Genet 95:66–72
R Development Core Team (2008). R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0. http://www.R-project.org
Uptmoor R, Wenzel W, Friedt W, Donaldson G, Ayisi K (2003) Comparative analysis on the genetic relatedness of sorghum bicolor accessions from Southern Africa by RADPs, AFLPs and SSRs. Theor Appl Genet 106:1316–1325
Weir BS (1996) Genetic data analysis II. Sinauer Associates, Sunderland
Acknowledgments
This study was funded by the United States Agency for International Development (USAID) Biotechnology and Biodversity Interface Program (BBI; Dr. Fabrice Sagnard), the Institute of plant Breeding and Population Genetics at the University of Hohenheim, Germany, and German Academic Exchange Service (DAAD: A0523923). USAID-BBI funded field experiments and collection trips. Costs of laboratory infrastructure and consumables at the Institute of plant Breeding and Population Genetics at the University of Hohenheim, Germany, were shared by BBI and the University of Hohenheim. We acknowledge the Kenya Agricultural Research Institute and Ben Kanyenji who supervised the collection of genetic materials in full compliance with regulations according to the Convention on Biological Diversity (CBD).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by X. Xia.
Electronic supplementary material
Below is the link to the electronic supplementary material.
122_2011_1608_MOESM4_ESM.doc
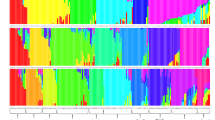
Figure S4: Bar plots of the INSTRUCT analysis. Each of the 62 wild sorghum populations is represented by a vertical bar being partitioned in up to K = 5 coloured segments that designate the population’s estimated membership fraction in the inferred subgroups. Populations are sorted according to regions of origin. Populations were sorted according to regions of origin and were collected from Turkana (1–17), Western (18–29), Coast (30–48) and Eastern (49–62) were collected regions of Kenya, respectively (DOC 369 kb)
Rights and permissions
About this article
Cite this article
Muraya, M.M., de Villiers, S., Parzies, H.K. et al. Genetic structure and diversity of wild sorghum populations (Sorghum spp.) from different eco-geographical regions of Kenya. Theor Appl Genet 123, 571–583 (2011). https://doi.org/10.1007/s00122-011-1608-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-011-1608-6