Summary
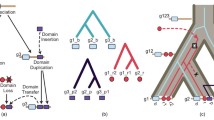
In the previous three reports in this series we demonstrated that the EF-hand family of proteins evolved by a complex pattern of gene duplication, transposition, and splicing. The dendrograms based on exon sequences are nearly identical to those based on protein sequences for troponin C, the essential light chain myosin, the regulatory light chain, and calpain. This validates both the computational methods and the dendrograms for these subfamilies. The proposal of congruence for calmodulin, troponin, C, essential light chain, and regulatory light chain was confirmed. There are, however, significant differences in the calmodulin dendrograms computed from DNA and from protein sequences.
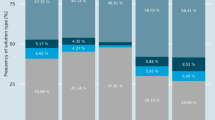
In this study we find that introns are distributed throughout the EF-hand domain and the interdomain regions. Further, dendrograms based on intron type and distribution bear little resemblance to those based on protein or on DNA sequences. We conclude that introns are inserted, and probably deleted, with relatively high frequency. Further, in the EF-hand family exons do not correspond to structural domains and exon shuffling played little if any role in the evolution of this widely distributed homolog family.
Calmodulin has had a turbulent evolution. Its dendrograms based on protein sequence, exon sequence, 3′-tail sequence, intron sequences, and intron positions all show significant differences.
Similar content being viewed by others
References
Banville D, Boie Y (1989) Retroviral long terminal repeat is the promoter of the gene encoding the tumor-associated calciumbinding protein oncomodulin in the rat. J Mol Biol 207:481–490
Barraclough R, Savin J, Dube SK, Rudland BS (1987) Molecular cloning and sequence of the gene for p9Ka. A cultured myoepithelial cell protein with strong homology to S-100, a calcium-binding protein. J Mol Biol 198:13–20
Barton PJR, Robert B, Cohen A, Garner I, Sassoon D, Weydert A, Buckingham ME (1988) Structure and sequence of the myosin alkali light chain gene expressed in adult cardiac atria and fetal striated muscle. J Biol Chem 263:12669–12676
Baum P, Furlong C, Byers B (1986) Yeast gene required for spindle pole body duplication: homology of its product with Ca2+-binding proteins. Proc Natl Acad Sci USA 83:5512–5516
Berchtold MW (1989) Parvalbumin genes from human and rat are identical in intron/exon organization and contain highly homologous regulatory elements and coding sequences. J Mol Biol 210:417–427
Berchtold MW (1993) Evolution of EF-hand calcium-modulated proteins. V. The genes encoding EF-hand proteins are not clustered in mammalian genomes. J Mol Evol 36:489–496
Berchtold MW, Epstein P, Beaudet AL, Payne ME, Heizmann CW, Means AR (1987) Structural organization and chromosomal assignment of the parvalbumin gene. J Biol Chem 262:8696–8701
Chung S-H, Swindle J (1990) Linkage of the calmodulin and ubiquitin loci inTrypanosoma cruzi. Nucleic Acids Res 18:4561–4569
Davis TN, Urdea MS, Masiarz FR, Thorner J (1986) Isolation of the yeast calmodulin gene: calmodulin is an essential protein. Cell 47:423–431
Deka N, Wong E, Matera AG, Kraft R, Leinwand LA, Schmid CW (1988) Repetitive nucleotide sequence insertions into a novel calmodulin-related gene and its processed pseudogene. Gene 71:123–134
Falkenthal S, Parker VP, Davidson N (1985) Developmental variations in the splicing pattern of transcripts from theDrosophila gene encoding myosin alkali light chain result in different carboxyl-terminal amino acid sequences. Proc Natl Acad Sci USA 82:449–453
Felsenstein J (1989) PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164–166
Feng D-F, Doolittle RF (1987) Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J Mol Evol 25:351–360
Ferrari S, Calabretta B, deRiel JK, Battini R, Ghezzo F, Lauret E, Griffin C, Emanuel BS, Gurrieri F, Baserga R (1987) Structural and functional analysis of a growth-regulated gene, the human calcyclin. J Biol Chem 262:8325–8332
Gahlmann R, Kedes L (1990) Cloning, structural analysis, and expression of the human fast twitch skeletal muscle troponin C gene. J Biol Chem 265:12520–12528
Gilbert W (1985) Genes-in-pieces revisited. Science 228:823–824
Gonzalez A, Lerner TJ, Huecas M, Sosa-Pineda B, Nogueira N, Lizardi PM (1985) Apparent generation of a segmented mRNA from two separate tandem gene families inTrypanosoma cruzi. Nucleic Acids Res 13:5789–5804
Grant JW, Zhong RQ, McEwen PM, Church SI (1990) Human nonsarcomeric 20,000 Da myosin regulatory light chain cDNA. Nucleic Acids Res 18:5892–5892
Hardin PE, Angerer LM, Hardin SH, Angerer RC, Klein WH (1988) Spec2 genes ofStrongylocentrotus purpuratus. Structure and differential expression in embryonic aboral ectoderm cells. J Mol Biol 202:417–431
Hardin SH, Carpenter CD, Hardin PE, Bruskin AM, Klein WH (1985) Structure of the Spec1 gene encoding a major calcium-binding protein in the embryonic ectoderm of the sea urchin,Strongylocentrotus purpuratus. J Mol Biol 186:243–255
Henderson SA, Spencer M, Sen A, Kumar C, Siddiqui MAQ, Chien KR (1989) Structure, organization, and expression of the rat cardiac myosin light chain-2 gene. Identification of a 250-base pair fragment which confers cardiac-specific expression. J Biol Chem 264:18142–18148
Karess RE, Chang Xj, Edwards KA, Kulkarni S, Aguilera I, Kiehart DP (1991) The regulatory light chain of nonmuscle myosin is encoded byspaghetti-squash, a gene required for cytokinesis inDrosophila. Cell 65:1177–1189
Kink JA, Maley ME, Preston RR, Ling K-Y, Wallen-Friedman MA, Saimi Y, Kung C (1990) Mutations inParamecium calmodulin indicate functional differences between the C-terminal and N-terminal lobesin vivo. Cell 62:165–174
Koller M, Schnyder B, Strehler EE (1990) Structural organization of the human CaMIII calmodulin gene. Biochim Biophys Acta 1087:180–189
Koller M, Baumer A, Strehler EE (1991) Characterization of two novel human retropseudogenes related to the calmodulinencoding gene, CaMII. Gene 97:245–251
Koller M, Strehler EE (1988) Characterization of an intronless human calmodulin-like pseudogene. FEBS Lett 239:121–128
Krisinger J, Darwish H, Maeda N, DeLuca HF (1988) Structure and nucleotide sequence of the rat intestinal vitamin D-dependent calcium binding protein gene. Proc Natl Acad Sci USA 85:8988–8992
Lee VD, Stapleton M, Huang B (1991) Genomic structure ofChlamydomonas caltractin. Evidence for intron insertion suggests a probable genealogy for the EF-hand superfamily of proteins. J Mol Biol 221:175–191
LéJohn HB (1989) Structure and expression of fungal calmodulin gene. J Biol Chem 264:19366–19372
Lenz S, Lohse P, Seidel U, Arnold H-H (1989) The alkali light chains of human smooth and nonmuscle myosins are encoded by a single gene. Tissue-specific expression by alternative splicing pathways. J Biol Chem 264:9009–9015
Miyake S, Emori Y, Suzuki K (1986) Gene organization of the small subunit of human calcium-activated neutral protease. Nucleic Acids Res 14:8805–8817
Moncrief ND, Kretsinger RH, Goodman M (1990) Evolution of EF-hand calcium-modulated proteins. I. Relationships based on amino acid sequences. J Mol Evol 30:522–562
Nabeshima Y-i, Nabeshima Y, Kawashima M, Nakamura S, Nonomura Y, Fujii-Kuriyarna Y (1988) Isolation of the chick myosin alkali light chain gene expressed in embryonic gizzard muscle and transitional expression of the light chain gene familyin vivo. J Mol Biol 204:497–505
Nabeshima Y-i, Fujii-Kuriyama Y, Muramatsu M, Ogata K (1984) Alternative transcription and two modes of splicing result in two myosin light chains from one gene. Nature 308:333–338
Nabeshima Y-i, Nabeshima Y, Kawashima M, Nakamura S, Nonomura Y, Fujii-Kuriyama Y (1988) Isolation of the chick myosin alkali light chain gene expressed in embryonic gizzard muscle and transitional expression of the light chain gene familyin vivo. J Mol Biol 204:497–505
Nakayama S, Kretsinger RH (1993) Evolution of EF-hand calcium-modulated proteins. III. Exon sequences confirm most dendrograms based on protein sequences. Calmodulin dendrograms show significant lack of parallelism. J Mol Evol 36:458–476
Nakayama S, Moncrief ND, Kretsinger RH (1992) Evolution of EF-hand calcium-modulated proteins. II. Domains of several subfamilies have diverse evolutionary histories. J Mol Evol 34:416–448
Nojima H (1989) Structural organization of multiple rat calmodulin genes. J Mol Biol 208:269–282
Nojima H, Sokabe H (1986) Structure of rat calmodulin processed genes with implications for a mRNA-mediated process of insertion. J Mol Biol 190:391–400
Nojima H, Sokabe H (1987) Structure of a gene for rat calmodulin. J Mol Biol 193:439–445
Nojima H, Kishi K, Sokabe H (1987) Multiple calmodulin mRNA species are derived from two distinct genes. Mol Cell Biol 7:1873–1880
Nudel U, Calvo JM, Shani M, Levy Z (1984) The nucleotide sequence of a rat myosin light chain 2 gene. Nucleic Acids Res 12:7175–7186
Ohno S, Emori Y, Imajoh S, Kawasaki H, Kisaragi M, Suzuki K (1984) Evolutionary origin of a calcium-dependent protease by fusion of genes for a thiol protease and a calcium-binding protein? Nature 312:566–570
Parker VP, Falkenthal S, Davidson N (1985) Characterization of the myosin light-chain-2 gene ofDrosophila melanogaster. Mol Cell Biol 5:3058–3068
Parmacek MS, Bengur AR, Vora AJ, Leiden JM (1990) The structure and regulation of expression of the murine fast skeletal troponin C gene. Identification of a developmentally regulated, muscle-specific transcriptional enhancer. J Biol Chem 265:15970–15976
Parmacek MS, Leiden JM (1989) Structure and expression of the murine slow/cardiac troponin C gene. J Biol Chem 264:13217–13225
Periasamy M, Wadgaonkar R, Kumar C, Martin BJ, Siddiqui MAQ (1989) Characterization of a rat myosin alkali light chain gene expressed in ventricular and slow twitch skeletal muscles. Nucleic Acids Res 17:7723–7735
Periasamy M, Strehler EE, Garfinkel LI, Gubits RM, Ruiz-Opazo N, Nadal-Ginard B (1984) Fast skeletal muscle myosin light chains 1 and 3 are produced from a single gene by a combined process of differential RNA transcription and splicing. J Biol Chem 259:13595–13604
Rasmussen CD, Means RL, Lu KP, May GS, Means AR (1990) Characterization and expression of the unique calmodulin gene ofAspergillus nidulans. J Biol Chem 265:13767–13775
Robert B, Daubas P, Akimenko M-A, Cohen A, Garner I, Guenet J-L, Buckingham M (1984) A single locus in the mouse encodes both myosin light chains 1 and 3, a second locus corresponds to a related pseudogene. Cell 39:129–140
Robson KJH, Jennings MW (1991) The structure of the calmodulin gene ofPlasmodium falciparum. Mol Biochem Parasitol 46:19–34
Salvato M, Sulston J, Albertson D, Brenner S (1986) A novel calmodulin-like gene from the nematodeCaenorhabditis elegans. J Mol Biol 190:281–290
Saporito SM, Sypherd PS (1991) The isolation and characterization of a calmodulin-encoding gene(CMDI) from the dimorphic fungusCandida albicans. Gene 106:43–49
Schreier T, Kedes L, Gahlmann R (1990) Cloning, structural analysis, and expression of the human slow twitch skeletal muscle/cardiac troponin C gene. J Biol Chem 265:21247–21253
Seharaseyon J, Bober E, Hsieh C-L, Fodor WL, Francke U, Arnold H-H, Vanin EF (1990) Human embryonic/atrial myosin alkali light chain gene: characterization, sequence, and chromosomal location. Genomics 7:289–293
SenGupta B, Detera-Wadleigh SD, McBride OW, Friedberg F (1989) A calmodulin pseudogene on human chromosome 17. Nucleic Acids Res 17:2868–2868
Smith VL, Doyle KE, Maune JF, Munjaal RP, Beckingham K (1990) Structure and sequence of theDrosophila melanogaster calmodulin gene. J Mol Biol 196:471–485
Stein JP, Munjaal RP, Lagacé L, Lai EC, O'Malley BW, Means AR (1983) Tissue-specific expression of a chicken calmodulin pseudogene lacking intervening sequences. Proc Natl Acad Sci USA 80:6485–6489
Strehler EE, Periasamy M, Strehler-Page M-A, Nadal-Ginard B (1985) Myosin light-chain 1 and 3 gene has two structurally distinct and differentially regulated promoters evolving at different rates. Mol Cell Biol 5:3168–3182
Swan DG, Cortes J, Hale RS, Leadlay PF (1989) Cloning, characterization, and heterologous expression of theSaccharopolyspora erythraea(Streptomvces erythraeus) gene encoding an EF-hand calcium-binding protein. J Bacteriol 171:5614–5619
Swanson ME, Sturner SF, Schwartz JH (1990) Structure and expression of theAplysia californica calmodulin gene. J Mol Biol 216:545–553
Takeda T, Yamamoto M (1987) Analysis and in vivo disruption of the gene coding for calmodulin inSchizosaccharomyces pombe. Proc Natl Acad Sci USA 84:3580–3584
Takemasa T, Ohnishi K, Kobayashi T, Takagi T, Konishi K, Watanabe Y (1989) Cloning and sequencing of the gene forTetrahymena calcium-binding 25-kDa protein (TCBP-25). J Biol Chem 264:19293–19301
Tschudi C, Young AS, Ruben L, Patton CL, Richards FF (1985) Calmodulin genes in trypanosomes are tandemly repeated and produce multiple mRNAs with a common 5′ leader sequence. Proc Natl Acad Sci USA 82:3998–4002
Tulchinsky EM, Grigorian MS, Ebralidze AK, Milshina NI, Lukanidin EM (1990) Structure of genemtsl, transcribed in metastatic mouse tumor cells. Gene 87:219–223
Wilson PW, Rogers J, Harding M, Pohl V, Pattyn G, Lawson DEM (1988) Structure of chick chromosomal genes for calbindin and calretinin. J Mol Biol 200:615–625
Wong S, Kretsinger RH, Campbell DA (1992) Identification of a new EF-hand superfamily member fromTrypanosoma brucei. Molec Gen Genet 233:225–230
Xiang M, Ge T, Tomlinson CR, Klein WH (1991) Structure and promoter activity of the LpS1 genes ofLytechinus pictus. Duplicated exons account for LpS1 proteins with eight calcium binding domains. J Biol Chem 266:10524–10533
Zimmer WE, Schloss JA, Silflow CD, Youngblom J, Watterson DM (1988) Structural organization, DNA sequence, and expression of the calmodulin gene. J Biol Chem 263:19370–19383
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Kretsinger, R.H., Nakayama, S. Evolution of EF-hand calcium-modulated proteins. IV. Exon shuffling did not determine the domain compositions of EF-hand Proteins. J Mol Evol 36, 477–488 (1993). https://doi.org/10.1007/BF02406723
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF02406723