Abstract
Stem rust, caused by Puccinia graminis f.sp. tritici (Pgt), is a destructive disease of wheat that has historically caused significant yield losses in much of the global wheat production area. Over the past 50 years, stem rust has been effectively controlled by deploying cultivars carrying stem rust resistance (Sr) genes. With the emergence of new Pgt races, namely Ug99 and its variants, there has been a reinvestment in stem rust research. This includes discovery, characterization, genetic mapping, and cloning of Sr genes. Here we investigated two such examples of genetic characterization and mapping of stem rust resistance. In the first example, a region on chromosome 6DS harbouring resistance to Ug99 was examined in several populations and from several sources. In the second example, a less typical genetic model of resistance was studied in which seedling resistance was activated by an independent locus exhibiting an apparent “nonsuppressing” effect. The knowledge gained by these and other lines of research will contribute to the goal of durable resistance to stem rust.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
Introduction
Stem rust, caused by Puccinia graminis Pers.:Pers. f. sp. tritici Eriks. & E. Henn. (Pgt), is a disease of wheat that has the potential to cause devastating losses in grain yield. The last epidemic of stem rust in North America occurred from 1953 to 1955 (Peturson 1958). Since then, resistant cultivars have successfully controlled the disease. However, the appearance of Pgt race TTKSK, also known as Ug99, rendered most of the globally grown wheat cultivars vulnerable to stem rust (Singh et al. 2011; Fetch et al. 2012). The distinguishing characteristic of this race is its virulence to Sr31, a broadly deployed stem rust resistance (Sr) gene (Pretorius et al. 2000). Furthermore, Ug99 has continued to evolve and accumulate different virulence combinations (Jin et al. 2009; Park et al. 2011). This has led to renewed efforts to find and deploy new Sr genes (e.g. Njau et al. 2010). More generally, this threat from Pgt has stimulated investment in several aspects of wheat, Pgt and their interactions. This has also underscored the need to employ multiple strategies to achieving resistance and utilise diverse resistance. To meet these needs it is critical to understand the resistance present in breeding material and cultivars, understand the relationship between different sources of resistance (e.g. allelism, linkage, breadth of resistance, target in the pathogen etc.), and develop tools to implement responsible breeding strategies.
It is in the context of renewed and diverse investigation into the wheat stem rust host-pathogen system that this paper was presented. Two lines of research were presented: (1) comparative mapping and fine-mapping of a chromosome region conferring qualitative resistance and (2) genetic interactions that enhance stem rust resistance at the seedling stage and in field conditions across multiple environments. Here, the pertinent literature for these studies is reviewed and our preliminary findings are discussed.
Comparative Mapping of Ug99 Resistance on Chromosome 6DS
SrCad
Most Canadian hexaploid cultivars are susceptible to Pgt races from the Ug99 race group and all of the cultivars that are grown on significant acreage are susceptible (Fetch et al. 2012). A screen of Canadian cultivars in Njoro, Kenya, where Ug99 races predominated, revealed that two hard red spring wheat cultivars, Peace and AC Cadillac, showed high levels of resistance. Peace and AC Cadillac are closely related cultivars and were believed to carry the same source of resistance to Ug99. Two populations were generated to study the inheritance and determine the chromosome location of the resistance (Hiebert et al. 2011). A doubled haploid (DH) population from the cross RL6071/Peace and an F2 population from the cross LMPG-6S/AC Cadillac both showed that a single gene was responsible for the Ug99 resistance detected at the seedling stage. Mapping with SSR markers showed that the resistance in both populations mapped distally on chromosome 6DS in the same genetic interval. A gene for common bunt resistance, Bt10, has been mapped to a similar chromosome region (Menzies et al. 2006). DNA marker FSD_RSA, a dominant marker that was developed to select Bt10 (Laroche et al. 2000), was added to the genetic maps of the Peace and AC Cadillac derived populations (Hiebert et al. 2011). FSD_RSA showed tight linkage to the Sr gene, which was given the temporary designation SrCad. Both AC Cadillac and Peace carry Bt10 which was inherited from the breeding line BW553. In the Canadian Prairie Spring (CPS) class of hexaploid wheat, BW553 was a parent to several cultivars and used as a donor of Bt10. A survey of CPS cultivars revealed that all of the cultivars that carried the positive allele (the allele found in BW553, Peace and AC Cadillac) were also resistant to Ug99 while all other cultivars were susceptible (Hiebert et al. 2011). In the germplasm screened with FSD_RSA, the positive allele is rare and has been linked to Ug99 resistance in every instance to date (Hiebert et al. 2011; Ghazvini et al. 2012). Thus, it appears that the presence of FSD_RSA is indicative of SrCad though it would be too large an assumption to deem the marker diagnostic as recombinants between FSD_RSA and SrCad were recovered in mapping populations (Hiebert et al. 2011; Hiebert et al. unpublished data). Interestingly, the CPS cultivars that carry SrCad showed a lower level resistance to Ug99 in field nurseries in Njoro, Kenya compared to Peace and AC Cadillac. Both Peace and AC Cadillac carry Lr34 while none of the CPS cultivars carrying SrCad carry Lr34. Hiebert et al. (2011) showed the combination of SrCad and Lr34 improved the level of Ug99 resistance in field conditions.
Sr42
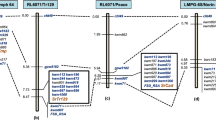
Given the map location of SrCad, there was interest in determining the relationship between SrCad and Sr42. Sr42 was reported to be carried on chromosome 6DS however there were no published genetic maps. Sr42 was found in Norin 40, a Japanese winter wheat (McIntosh et al. 1995). Using a DH population from the cross LMPG-6S/Norin 40, Ghazvini et al. (2012) mapped Sr42 using SSR markers and FSD_RSA. Sr42 mapped to the same genetic interval as SrCad. Norin 40, like Peace and AC Cadillac, carries the positive allele of FSD_RSA. Given that this marker allele is rare, it seems likely that Norin 40 and BW533, the donor of SrCad to all Canadian carriers, have a parent in common however no such parent could be traced. The sizes of the DH populations used to map SrCad (n = 295; Hiebert et al. 2011) and Sr42 (n = 248; Ghazvini et al. 2012) yielded good map resolution and their respective map positions are not significantly different (Fig. 20.1). Both SrCad and Sr42 conferred resistance to Pgt races Ug99 and RTQSC. While very limited, this does show an additional similarity between these resistances. New markers developed in ongoing fine-mapping experiments were unable to show differences in map position between SrCad and Sr42 (unpublished data). It is likely that SrCad and Sr42 are allelic and probably represent the same allele.
Other Sr Genes on Chromosome 6DS that Confer Resistance to Ug99 Stem Rust
There have other Sr genes mapped to 6DS in a similar chromosome region as SrCad and Sr42 that confer resistance to Ug99. One such example was reported by Olsen et al. (2013) where an Sr gene, temporarily designated SrTA10187, identified in Aegilops tauschii was transferred to common wheat and was mapped to chromosome 6DS. In that study, few DNA markers were mapped, however two of the three markers were also used to map SrCad (Hiebert et al. 2011). The map positions of SrCad and SrTA10187 cannot be differentiated based on the data published to date. Recently, seedling resistance to Ug99 was mapped in five CIMMYT spring wheat cultivars and one US winter wheat cultivar (Lopez-Vera et al. 2014). In all six populations the resistance mapped to a similar region of chromosome 6DS however the map order varied slightly between the various maps. Whether the map differences are the result of experimental error such as misclassified progeny, or the detection of different genes is unknown presently. The authors postulate that two different genes were mapped, one that is near or allelic to SrCad/Sr42 and one that is distal to these genes but closely linked. It was suggested that the resistance postulated to be allelic to SrCad/Sr42 was SrTmp derived from the cultivar Triumph 64 and our preliminary map of SrTmp that would agree with this assertion. While Lopez-Vera et al. (2014) are basing their hypothesis on gene postulation experiments, we used a DH population from the cross LMPG-6S/Triumph 64 to generate a population to directly determine the map position of SrTmp. Using the LMPG-6S/Norin 40 and LMPG-6S/Triumph 64 DH populations we were able to show that Sr42 and SrTmp differ in the breadth of resistance they confer. Thus, even if the two are allelic, they represent different allelic forms.
Conclusion
Resistance to Ug99-type stem rust races has been mapped to chromosome 6DS in a number of studies from a number of sources. Understanding the relationships between these genes is important for developing strategies to deploy these genes. If the genes are allelic it is prudent to determine which allele is the most relevant for a given growing region and the local Pgt population. If the genes are non-allelic it could be worthwhile to attempt to recover recombinants carrying gene combinations to generate a gene stack carried in a tight linkage group. The DNA markers developed from the fine-mapping of SrCad (unpublished) could be useful in delineating the Sr genes mapped to this chromosome region. To further characterize these genes phenotypically, near-isogenic lines (NILs) should be produced to allow an accurate comparison of their range of effectiveness against a panel of Pgt races.
Chromosome 7D: Carrier of a Suppressor and a Nonsuppressor
A Suppressor of Stem Rust Resistance
Canthatch is an old Canadian hard red spring wheat cultivar that is a derivative of Thatcher (McCallum and DePauw 2008). Canthatch is susceptible to several races of Pgt and was used in many genetic studies by Dr. Eric Kerber (retired, AAFC, Winnipeg, Canada). In an early experiment, the A and B genome component of Canthatch was isolated to generate an ‘extracted tetraploid’ which had the genome constitution of 2n = 4x, AABB (Kerber 1964). The extracted tetraploid, named Tetra Canthatch, lacked vigour, had low fertility, and was morphologically different from Canthatch and common tetraploids. Furthermore, Tetra Canthatch had seedling resistance to some Pgt races that were virulent to Canthatch. This implied that the D genome or a component of the D genome was suppressing Sr genes present on the A and/or B genomes in Canthatch. Using nullisomic and ditelosomic stocks in a Canthatch background, it was demonstrated that the suppression of Sr genes was caused by the long arm of chromosome 7D (Kerber and Green 1980). Induced mutants showed the same phenotype as Tetra Canthatch and Canthatch ditelo 7DS. Furthermore, the suppression was caused by a single gene on chromosome 7DL (Kerber 1991). Telocentric mapping showed that the suppressor locus was independent of the centromere.
Adult-Plant Resistance Genes Can Act as a Nonsuppressor
Lr34 is a well-known adult-plant resistance (APR) gene that confers quantitative resistance to all of three of the rust disease of wheat and is located on the short arm of chromosome 7D (Dyck 1987; Singh et al. 2012). In addition to conferring APR, Lr34 was associated with nonsuppression of seedling stem rust resistance in a Thatcher background (Dyck 1987; Kerber and Aung 1999). Comparing Canthatch, Thatcher, Tetra Canthatch, Canthatch-nulli 7D and RL6058 (a Thatcher NIL carrying Lr34) with four races of Pgt, Dyck (1987) showed that (1) Canthatch and Thatcher were susceptible to all four races of Pgt, (2) Tetra Canthatch and Canthatch-nulli 7D were resistant to three of these races, (3) RL6058 was resistant to the same three races as Tetra Canthatch and Canthatch-nulli 7D, and (4) one race was virulent on all five genotypes. Thus, it appears that Lr34 negates the effect of the suppressor locus and that that the stem rust resistance expressed in Thatcher-type wheats by either removing the suppressor or adding Lr34 is race-specific. Figure 20.2 shows a conceptual drawing of the relationship between the suppressor locus and Lr34, although the actual mechanisms are not presently understood.
A conceptual summary of the interaction of the suppressor (Sup) and Lr34 on chromosome 7D. (a) The suppressor prevents the expression of seedling Sr genes from the A and/or B genomes of Thatcher-type wheats if the susceptible allele of Lr34 is present. (b) Knocking out the suppressor by mutagenesis allows the expression of the otherwise suppressed Sr genes. (c) The resistant allele of Lr34 appears to negate the effect of the suppressor and allows the expression of the Sr genes
Another Thatcher NIL, RL6077, showed the same pattern of seedling stem rust resistance as RL6058 (Dyck 1987). RL6077 carries the gene Lr67 which is similar to Lr34 in its phenotype and its multiple-disease resistance (Hiebert et al. 2010). A preliminary study showed that Lr67 was responsible for the expression of seedling stem rust resistance in RL6077 and appeared to act as a nonsuppressor like Lr34 (Hiebert et al. 2012). Thus, two unique APR genes, which both confer multiple pest resistance, also appear to act as nonsuppressors of seedling stem rust resistance genes found in Thatcher-type wheats.
We presented preliminary data in three areas to (1) assess Lr34 as a nonsuppressor, (2) preliminary map position of seedling stem rust resistance in Thatcher expressed in the presence of Lr34, and (3) preliminary map position of stem rust resistance in the field that is expressed in the presence of Lr34. Using mutants of Lr34 in RL6058 (Spielmeyer et al. 2013), it appears that mutant lines that have lost Lr34 activity have also lost the seedling stem rust resistance observed in the presence of a normal Lr34 allele. Thus, we tentatively claim that it is Lr34 that is responsible for the nonsupressing character observed in RL6058 and not a gene tightly linked to Lr34. We developed two populations that are fixed for Lr34 but segregate for Thatcher-derived seedling stem rust resistance and resistance in the field. Seedling resistance and field resistance were mapped as QTL in these preliminary analyses and chromosome 3B significantly contributed to both traits. These preliminary findings need to be confirmed and experiments are ongoing to do so.
Conclusions
One aspect of the relationship between the suppressor on 7DL and Lr34 that is presently unknown is whether removing or inactivating the suppressor leads to the expression of the same Sr genes that are expressed in the presence of Lr34. Similarly, we do not know if Lr67 and Lr34 are interacting with the same genes. These lines of investigation are ongoing and should fill in the gaps in our knowledge of these interactions. From a practical standpoint, understanding these genetic interactions could be important for wheat breeders. It would easier to retain Sr genes (i.e. the suppressed genes) in elite breeding material and add, for example, Lr34 compared to the effort of introducing new or different Sr genes to enhance to stem rust resistance in new cultivars. In Canada, Thatcher comprises a large component of coefficient of parentage for most of the widely grown hard red spring wheat cultivars (McCallum and DePauw 2008). Thus, identifying the genes in Thatcher that are expressed in the presence of Lr34 is directly applicable.
Concluding Remarks
An array of strategies can be employed to achieve resistance to stem rust. There is not a “one size fits all” approach that must be followed. However, genetic resources for resistance to stem rust are valuable and implementing a strategy that prolongs the usefulness of Sr genes is the responsible approach. In order to accomplish this we must understand which genes are present in our germplasm, determine the relationships between genes and their breadths of effectiveness, develop tools for molecular breeding approaches, and unravel the genetic interactions that can lead us to more durable resistance. Here we examined two examples of the lines of research that can help geneticists, pathologist and breeders reach the goal of effective and durable resistance. With the recent investment in various aspects of wheat stem rust research more tools and resources are becoming available. It is now our responsibility to translate the advances made in the lab into rust resistant wheat cultivars in the field.
References
Dyck PL (1987) The association of a gene for leaf rust resistance with the chromosome 7D suppressor of stem rust resistance in common wheat. Genome 29:467–469
Fetch TG Jr, Zegeye T, Singh D et al (2012) Virulence of Ug99 (race TTKSK) and race TRTTF on Canadian wheat cultivars. Can J Plant Sci 92:602
Ghazvini H, Hiebert CW, Zegeye T et al (2012) Inheritance of resistance to Ug99 stem rust in wheat cultivar Norin 40 and genetic mapping of Sr42. Theor Appl Genet 125:817–824
Hiebert CW, Thomas JB, McCallum BD et al (2010) An introgression on wheat chromosome 4DL in RL6077 (Thatcher*6/PI 250413) confers adult plant resistance to stripe rust and leaf rust (Lr67). Theor Appl Genet 121:1083–1091
Hiebert CW, Fetch TG, Zegeye T et al (2011) Genetics and mapping of seedling resistance to Ug99 stem rust in Canadian wheat cultivars ‘Peace’ and ‘AC Cadillac’. Theor Appl Genet 122:143–149
Hiebert C, Fetch T, McCallum B (2012) Association of Lr67 with nonsuppression of seedling stem rust resistance in the wheat cultivar Thatcher. In: 13th international cereal rust and powdery mildew conference, Beijing
Jin Y, Szabo L, Rouse M et al (2009) Detection of virulence to resistance gene Sr36 within race TTKS lineage of Puccinia graminis f. sp. tritici. Plant Dis 93:367–370
Kerber ER (1964) Wheat: reconstitution of the tetraploid component (AABB) of hexaploids. Science 143:253–255
Kerber ER (1991) Stem-rust resistance in ‘Canthatch’ hexaploid wheat induced by a nonsuppressor mutation on chromosome 7DL. Genome 34:935–939
Kerber ER, Aung T (1999) Leaf rust resistance gene Lr34 associated with nonsuppression of stem rust resistance in the wheat cultivar Canthatch. Phytopathology 89:518–521
Kerber ER, Green GJ (1980) Suppression of stem rust resistance in the hexaploid wheat cv. Canthatch by chromosome 7DL. Can J Bot 58:1347–1350
Laroche A, Demeke T, Gaudet DA et al (2000) Development of a PCR marker for rapid identification of the Bt-10 gene for common bunt resistance in wheat. Genome 43:217–223
Lopez-Vera EE, Nelson S, Singh RP et al (2014) Resistance to stem rust Ug99 in six bread wheat cultivars maps to chromosome 6DS. Theor Appl Genet 127:231–239
McCallum BD, DePauw RM (2008) A review of wheat cultivars grown in the Canadian prairies. Can J Plant Sci 88:649–677
McIntosh RA, Wellings CR, Park RF (1995) Wheat rusts: an atlas of resistance genes. CSIRO Publications, East Melbourne, pp 85–141
Menzies JG, Knox RE, Popovic Z, Procunier JD (2006) Common bunt resistance gene Bt10 located on wheat chromosome 6D. Can J Plant Sci 86:1409–1412
Njau PN, Jin Y, Huerta-Espino J et al (2010) Identification and evaluation of sources of resistance to stem rust race Ug99 in wheat. Plant Dis 94:413–419
Olsen EL, Rouse MN, Pumphrey MO et al (2013) Introgression of stem rust resistance genes SrTA10187 and SrTA10171 from Aegilops tauschii to wheat. Theor Appl Genet 126:2477–2484
Park R, Fetch T, Hodson D et al (2011) International surveillance of wheat rust pathogen: progress and challenges. Euphytica 179:109–117
Peturson B (1958) Wheat rust epidemics in western Canada in 1953, 1954, and 1955. Can J Plant Sci 38:16–28
Pretorius ZA, Singh RP, Wagoire WW, Payne TS (2000) Detection of virulence to wheat stem rust resistance gene Sr31 in Puccinia graminis f. sp. tritici in Uganda. Plant Dis 84:203
Singh RP, Hodson DP, Huerta-Espino J et al (2011) The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Annu Rev Phytopathol 49:465–481
Singh RP, Herrera-Foessel SA, Huerta-Espino J et al (2012) Lr34/Yr18/Sr57/Pm38/Bdv1/Ltn1 confers slow rusting, adult plant resistance to Puccinia graminis tritici. 2012 Borlaug global rust initiative technical workshop, Beijing
Spielmeyer W, Mago R, Wellings C, Ayliffe M (2013) Lr67 and Lr34 rust resistance genes have much in common – they confer broad spectrum resistance to multiple pathogens in wheat. BMC Plant Biol 13:96
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is distributed under the terms of the Creative Commons Attribution Noncommercial License, which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Copyright information
© 2015 Her Majesty the Queen in Right of Canada
About this paper
Cite this paper
Hiebert, C. et al. (2015). Stem Rust Resistance: Two Approaches. In: Ogihara, Y., Takumi, S., Handa, H. (eds) Advances in Wheat Genetics: From Genome to Field. Springer, Tokyo. https://doi.org/10.1007/978-4-431-55675-6_20
Download citation
DOI: https://doi.org/10.1007/978-4-431-55675-6_20
Publisher Name: Springer, Tokyo
Print ISBN: 978-4-431-55674-9
Online ISBN: 978-4-431-55675-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)