Abstract
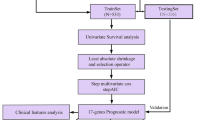
Breast cancer is the most prevalent cancer, and it is accompanied by high heterogeneity. N6-methyladenosine (m6A) modification significantly contributes to breast cancer tumorigenesis and progression. However, how m6A-related genes affect the clinical outcomes and tumor immune microenvironment (TIME) of breast cancer is largely unknown. Our study developed an m6A-related gene signature on the basis of The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases. The m6A-related gene signature was constructed using univariate, least absolute shrinkage and selection operator (LASSO), and multivariate Cox regression analyses. Breast cancer patients were classified into low- and high-risk groups depending on the median risk score. The reliability and efficiency of the signature were validated using Kaplan–Meier analysis, receiver operating characteristic (ROC) curves, and principal component analysis (PCA). The risk score was validated as an independent indicator associated with overall survival, and a nomogram model was created to estimate the overall survival of patients with breast cancer. Functional annotation suggested that the risk score had a strong relationship with immune-related pathways. Different proportions of immune cell infiltration between the two groups were evaluated using various algorithms. The high-risk group had higher immune checkpoint expression levels. We discovered that one of the 6 prognostic genes, TMEM71, was downregulated in breast cancer tissues. In vitro experiments indicated that overexpression of TMEM71 suppressed breast cancer cell proliferation and migration. In conclusion, the m6A-related gene signature may be a sensitive biomarker for overall survival prediction and guide the individualized treatment for breast cancer patients.








Similar content being viewed by others
Data availability statement
The RNA-seq data and clinical data of breast cancer used for the training and validation cohorts were downloaded from the UCSC Cancer Browser (https://xenabrowser.net/datapages/) and GEO (https://www.ncbi.nlm.nih.gov/gds/). The experimental datasets used and analyzed in this study are available from the corresponding authors upon reasonable request.
Abbreviations
- AJCC:
-
The American Joint Committee on Cancer
- AUC:
-
Areas under curve
- DC:
-
Dendritic cell
- DEG:
-
Differentially expressed gene
- EMT:
-
Epithelial–mesenchymal transition
- GEO:
-
Gene Expression Omnibus
- GO:
-
Gene Ontology
- GSEA:
-
Gene Set Enrichment Analysis
- HUC:
-
Human urothelial carcinoma
- ICB:
-
Immune checkpoint blockade
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- LASSO:
-
Least absolute shrinkage and selection operator
- m6A:
-
N6-methyladenosine
- NSCLC:
-
Non-small cell lung cancer
- PCA:
-
Principal component analysis
- PRCC:
-
Papillary renal cell carcinoma
- PVDF:
-
Polyvinylidene fluoride
- RIPA:
-
Radio immunoprecipitation assay
- ROC:
-
Receiver operating characteristic
- ssGSEA:
-
Single-sample gene set enrichment analysis
- TAM:
-
Tumor-associated macrophages
- TCGA:
-
The Cancer Genome Atlas
- TIME:
-
Tumor immune microenvironment
- TMEM:
-
Transmembrane
References
Siegel RL, Miller KD, Fuchs HE, Jemal A. 2022 Cancer statistics. CA Cancer J Clin. 2022;72(1):7–33. https://doi.org/10.3322/caac.21708.
Kroenke CH, Michael YL, Poole EM, Kwan ML, Nechuta S, Leas E, et al. Postdiagnosis social networks and breast cancer mortality in the after breast cancer pooling project. Cancer. 2017;123(7):1228–37. https://doi.org/10.1002/cncr.30440.
Akira S, Maeda K. Control of RNA stability in immunity. Annu Rev Immunol. 2021;39:481–509. https://doi.org/10.1146/annurev-immunol-101819-075147.
Chen XY, Zhang J, Zhu JS. The role of m(6)A RNA methylation in human cancer. Mol Cancer. 2019;18(1):103. https://doi.org/10.1186/s12943-019-1033-z.
Huang H, Weng H, Chen J. m(6)A Modification in coding and non-coding RNAs: roles and therapeutic implications in cancer. Cancer Cell. 2020;37(3):270–88. https://doi.org/10.1016/j.ccell.2020.02.004.
Wei M, Bai JW, Niu L, Zhang YQ, Chen HY, Zhang GJ. The complex roles and therapeutic implications of m(6)A modifications in breast cancer. Front Cell Develop Biol. 2020;8: 615071. https://doi.org/10.3389/fcell.2020.615071.
Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun L, et al. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol Cancer. 2019;18(1):46. https://doi.org/10.1186/s12943-019-1004-4.
Chen F, Chen Z, Guan T, Zhou Y, Ge L, Zhang H, et al. N(6) -Methyladenosine regulates mRNA stability and translation efficiency of KRT7 to promote breast cancer lung metastasis. Can Res. 2021;81(11):2847–60. https://doi.org/10.1158/0008-5472.Can-20-3779.
Li B, Jiang J, Assaraf YG, Xiao H, Chen ZS, Huang C. Surmounting cancer drug resistance: New insights from the perspective of N(6)-methyladenosine RNA modification. Drug Resistance Updates Rev Comment Antimicrob Anticancer Chemother. 2020;53: 100720. https://doi.org/10.1016/j.drup.2020.100720.
Chen J, Jiang CC, Jin L, Zhang XD. Regulation of PD-L1: a novel role of pro-survival signalling in cancer. Ann Oncol Off J Eur Soc Med Oncol. 2016;27(3):409–16. https://doi.org/10.1093/annonc/mdv615.
Wan W, Ao X, Chen Q, Yu Y, Ao L, Xing W, et al. METTL3/IGF2BP3 axis inhibits tumor immune surveillance by upregulating N(6)-methyladenosine modification of PD-L1 mRNA in breast cancer. Mol Cancer. 2022;21(1):60. https://doi.org/10.1186/s12943-021-01447-y.
Zhang Z, Zhang C, Luo Y, Zhang G, Wu P, Sun N, et al. RNA N(6) -methyladenosine modification in the lethal teamwork of cancer stem cells and the tumor immune microenvironment current landscape and therapeutic potential. Clin Transl Med. 2021;11(9): e525. https://doi.org/10.1002/ctm2.525.
Dong L, Chen C, Zhang Y, Guo P, Wang Z, Li J, et al. The loss of RNA N(6)-adenosine methyltransferase Mettl14 in tumor-associated macrophages promotes CD8(+) T cell dysfunction and tumor growth. Cancer Cell. 2021;39(7):945-57.e10. https://doi.org/10.1016/j.ccell.2021.04.016.
Yu X, Dong P, Yan Y, Liu F, Wang H, Lv Y, et al. Identification of N6-methyladenosine-associated long non-coding RNAs for immunotherapeutic response and prognosis in patients with pancreatic cancer. Front cell Develop Biol. 2021;9: 748442. https://doi.org/10.3389/fcell.2021.748442.
Zhu W, Ye Z, Chen L, Liang H, Cai Q. A pyroptosis-related lncRNA signature predicts prognosis and immune microenvironment in head and neck squamous cell carcinoma. Int Immunopharmacol. 2021;101(Pt B): 108268. https://doi.org/10.1016/j.intimp.2021.108268.
Curigliano G, Burstein HJ, Winer EP, Gnant M, Dubsky P, Loibl S et al. 2017 De-escalating and escalating treatments for early-stage breast cancer: the St Gallen International expert consensus conference on the primary therapy of early breast cancer 2017. An Oncol Off J Eur Soc Med Oncol https://doi.org/10.1093/annonc/mdx308.
Xu Q, Yan X, Han Z, Jin X, Jin Y, Sun H, et al. Immune cell infiltration and relevant gene signatures in the tumor microenvironment that significantly associates with the prognosis of patients with breast cancer. Front Mol Biosci. 2022;9: 823911. https://doi.org/10.3389/fmolb.2022.823911.
Kwa M, Makris A, Esteva FJ. Clinical utility of gene-expression signatures in early stage breast cancer. Nat Rev Clin Oncol. 2017;14(10):595–610. https://doi.org/10.1038/nrclinonc.2017.74.
Subramanian J, Simon R. Gene expression-based prognostic signatures in lung cancer: ready for clinical use? J Natl Cancer Inst. 2010;102(7):464–74. https://doi.org/10.1093/jnci/djq025.
Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019;20(10):608–24. https://doi.org/10.1038/s41580-019-0168-5.
Jiao J, Jiang L, Luo Y. N6-methyladenosine-related RNA signature predicting the prognosis of ovarian cancer. Recent Pat Anti-Cancer Drug Discovery. 2021;16(3):407–16. https://doi.org/10.2174/1574892816666210615164645.
Na Z, Fan L, Wang X. Gene signatures and prognostic values of n6-methyladenosine related genes in ovarian cancer. Front Genet. 2021;12: 542457. https://doi.org/10.3389/fgene.2021.542457.
Zhang L, Luo Y, Cheng T, Chen J, Yang H, Wen X, et al. Development and validation of a prognostic n6-methyladenosine-related immune gene signature for lung adenocarcinoma. Pharmacogenom Personal Med. 2021;14:1549–63. https://doi.org/10.2147/pgpm.S332683.
Jiang D, LaGory EL, Kenzelmann Brož D, Bieging KT, Brady CA, Link N, et al. Analysis of p53 transactivation domain mutants reveals Acad11 as a metabolic target important for p53 pro-survival function. Cell Rep. 2015;10(7):1096–109. https://doi.org/10.1016/j.celrep.2015.01.043.
Xie Q, Xiao YS, Jia SC, Zheng JX, Du ZC, Chen YC, et al. FABP7 is a potential biomarker to predict response to neoadjuvant chemotherapy for breast cancer. Cancer Cell Int. 2020;20(1):562. https://doi.org/10.1186/s12935-020-01656-3.
Zhang Y, Di X, Chen G, Liu J, Zhang B, Feng L, et al. An immune-related signature that to improve prognosis prediction of breast cancer. Am J Cancer Res. 2021;11(4):1267–85.
Shen CH, Wu JY, Wang SC, Wang CH, Hong CT, Liu PY, et al. The suppressive role of phytochemical-induced glutathione S-transferase Mu 2 in human urothelial carcinoma cells. Biomed pharmacother. 2022. https://doi.org/10.1016/j.biopha.2022.113102.
Tang SC, Wu CH, Lai CH, Sung WW, Yang WJ, Tang LC, et al. Glutathione S-transferase mu2 suppresses cancer cell metastasis in non-small cell lung cancer. Mol Cancer Res MCR. 2013;11(5):518–29. https://doi.org/10.1158/1541-7786.Mcr-12-0488.
Yang M, Zhang T, Zhang Y, Ma X, Han J, Zeng K, et al. MYLK4 promotes tumor progression through the activation of epidermal growth factor receptor signaling in osteosarcoma. J Exp Clin Cancer Res CR. 2021;40(1):166. https://doi.org/10.1186/s13046-021-01965-z.
Marx S, Dal Maso T, Chen JW, Bury M, Wouters J, Michiels C, et al. Transmembrane (TMEM) protein family members: Poorly characterized even if essential for the metastatic process. Semin Cancer Biol. 2020;60:96–106. https://doi.org/10.1016/j.semcancer.2019.08.018.
Lee H, Goodarzi H, Tavazoie SF, Alarcón CR. TMEM2 Is a SOX4-regulated gene that mediates metastatic migration and invasion in breast cancer. Can Res. 2016;76(17):4994–5005. https://doi.org/10.1158/0008-5472.Can-15-2322.
Zhu H, Su Z, Ning J, Zhou L, Tan L, Sayed S, et al. Transmembrane protein 97 exhibits oncogenic properties via enhancing LRP6-mediated Wnt signaling in breast cancer. Cell Death Dis. 2021;12(10):912. https://doi.org/10.1038/s41419-021-04211-8.
Li M, Han Y, Zhou H, Li X, Lin C, Zhang E, et al. Transmembrane protein 170B is a novel breast tumorigenesis suppressor gene that inhibits the Wnt/β-catenin pathway. Cell Death Dis. 2018;9(2):91. https://doi.org/10.1038/s41419-017-0128-y.
Liu Z, Wan Y, Yang M, Qi X, Dong Z, Huang J, et al. Identification of methylation-driven genes related to the prognosis of papillary renal cell carcinoma: a study based on the cancer genome atlas. Cancer Cell Int. 2020;20:235. https://doi.org/10.1186/s12935-020-01331-7.
Ythier D, Larrieu D, Binet R, Binda O, Brambilla C, Gazzeri S, et al. Sumoylation of ING2 regulates the transcription mediated by Sin3A. Oncogene. 2010;29(44):5946–56. https://doi.org/10.1038/onc.2010.325.
Bai F, Jin Y, Zhang P, Chen H, Fu Y, Zhang M, et al. Bioinformatic profiling of prognosis-related genes in the breast cancer immune microenvironment. Aging. 2019;11(21):9328–47. https://doi.org/10.18632/aging.102373.
Costa A, Kieffer Y, Scholer-Dahirel A, Pelon F, Bourachot B, Cardon M, et al. Fibroblast heterogeneity and immunosuppressive environment in human breast cancer. Cancer Cell. 2018;33(3):463-79.e10. https://doi.org/10.1016/j.ccell.2018.01.011.
Savas P, Salgado R, Denkert C, Sotiriou C, Darcy PK, Smyth MJ, et al. Clinical relevance of host immunity in breast cancer: from TILs to the clinic. Nat Rev Clin Oncol. 2016;13(4):228–41. https://doi.org/10.1038/nrclinonc.2015.215.
Germain RN. T-cell development and the CD4-CD8 lineage decision. Nat Rev Immunol. 2002;2(5):309–22. https://doi.org/10.1038/nri798.
Kishton RJ, Sukumar M, Restifo NP. Metabolic regulation of t cell longevity and function in tumor immunotherapy. Cell Metab. 2017;26(1):94–109. https://doi.org/10.1016/j.cmet.2017.06.016.
Gardner A, Ruffell B. Dendritic cells and cancer immunity. Trends Immunol. 2016;37(12):855–65. https://doi.org/10.1016/j.it.2016.09.006.
Shaul ME, Fridlender ZG. Tumour-associated neutrophils in patients with cancer. Nat Rev Clin Oncol. 2019;16(10):601–20. https://doi.org/10.1038/s41571-019-0222-4.
Li W, Wang X, Li C, Chen T, Yang Q. Exosomal non-coding RNAs: Emerging roles in bilateral communication between cancer cells and macrophages. Mol Ther J Am Soc Gene Ther. 2022;30(3):1036–53. https://doi.org/10.1016/j.ymthe.2021.12.002.
Zhang J, Shan B, Lin L, Dong J, Sun Q, Zhou Q, et al. Dissecting the role of N6-methylandenosine-related long non-coding RNAs signature in prognosis and immune microenvironment of breast cancer. Front cell Develop Biol. 2021;9: 711859. https://doi.org/10.3389/fcell.2021.711859.
Acknowledgements
Not applicable
Funding
This work was supported by National Key Research and Development Program (No. 2020YFA0712400), Special Foundation for Taishan Scholars (No. ts20190971), National Natural Science Foundation of China (No. 81874119; No. 82072912; No. 82004122), China Postdoctoral Science Foundation (No. 2020M682199), Shandong Provincial Natural Science Foundation, China (No. ZR2020QH335, No. ZR2019LZL003), Chen Xiao-ping Foundation for the Development of Science and Technology of Hubei Province (CXPJJH121001-2021003), 2021 Shandong Medical Association Clinical Research Fund—Qilu Special Project (YXH2022ZX02160), Foundation from Clinical Research Center of Shandong University (No.2020SDUCRCA015), Qilu Hospital Clinical New Technology Developing Foundation (No. 2019-3).
Author information
Authors and Affiliations
Contributions
Conceived and designed the study: WL and XW. Data collection: CL. Data analysis: ZL. Generated figures: TC. Wrote the manuscript: XZ. Revised and edited the manuscript: QY. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Ethical approval
The study was approved by the Ethics Committee on Scientific Research of Shandong University, Qilu Hospital (approval number: KYLL-2016-350), and informed consents were provided by all patients.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, W., Wang, X., Li, C. et al. Identification and validation of an m6A-related gene signature to predict prognosis and evaluate immune features of breast cancer. Human Cell 36, 393–408 (2023). https://doi.org/10.1007/s13577-022-00826-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-022-00826-x