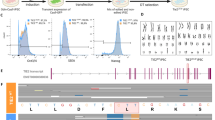
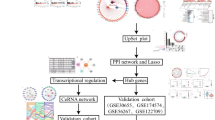
Abstract
Patients with familial cerebral cavernous malformation (CCM) inherit germline loss of function mutations and are susceptible to progressive development of brain lesions and neurological sequelae during their lifetime. To date, no homologous circulating molecules have been identified that can reflect the presence of germ line pathogenetic CCM mutations, either in animal models or patients. We hypothesize that homologous differentially expressed (DE) plasma miRNAs can reflect the CCM germline mutation in preclinical murine models and patients. Herein, homologous DE plasma miRNAs with mechanistic putative gene targets within the transcriptome of preclinical and human CCM lesions were identified. Several of these gene targets were additionally found to be associated with CCM-enriched pathways identified using the Kyoto Encyclopedia of Genes and Genomes. DE miRNAs were also identified in familial-CCM patients who developed new brain lesions within the year following blood sample collection. The miRNome results were then validated in an independent cohort of human subjects with real-time-qPCR quantification, a technique facilitating plasma assays. Finally, a Bayesian-informed machine learning approach showed that a combination of plasma levels of miRNAs and circulating proteins improves the association with familial-CCM disease in human subjects to 95% accuracy. These findings act as an important proof of concept for the future development of translatable circulating biomarkers to be tested in preclinical studies and human trials aimed at monitoring and restoring gene function in CCM and other diseases.





Similar content being viewed by others
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Code Availability
Not applicable.
References
Awad IA, Polster SP. Cavernous angiomas: deconstructing a neurosurgical disease. J Neurosurg. 2019;131:1–13. https://doi.org/10.3171/2019.3.JNS181724.
Akers A, Al-Shahi Salman R, I AA, Dahlem K, Flemming K, Hart B, et al. Synopsis of guidelines for the clinical management of cerebral cavernous malformations: consensus recommendations based on systematic literature review by the Angioma Alliance scientific advisory board clinical experts panel. Neurosurgery. 2017;80:665–80. https://doi.org/10.1093/neuros/nyx091.
Shenkar R, Shi C, Rebeiz T, Stockton RA, McDonald DA, Mikati AG, et al. Exceptional aggressiveness of cerebral cavernous malformation disease associated with PDCD10 mutations. Genet Med. 2015;17:188–96. https://doi.org/10.1038/gim.2014.97.
Polster SP, Cao Y, Carroll T, Flemming K, Girard R, Hanley D, et al. Trial readiness in cavernous angiomas with symptomatic hemorrhage (CASH). Neurosurgery. 2019;84:954–64. https://doi.org/10.1093/neuros/nyy108.
Snellings DA, Hong CC, Ren AA, Lopez-Ramirez MA, Girard R, Srinath A, et al. Cerebral cavernous malformation: from mechanism to therapy. Circ Res. 2021;129:195–215. https://doi.org/10.1161/CIRCRESAHA.121.318174.
Zhu W, Shen F, Mao L, Zhan L, Kang S, Sun Z, et al. Soluble FLT1 gene therapy alleviates brain arteriovenous malformation severity. Stroke. 2017;48:1420–3. https://doi.org/10.1161/STROKEAHA.116.015713.
Girard R, Li Y, Stadnik A, Shenkar R, Hobson N, Romanos S, et al. A roadmap for developing plasma diagnostic and prognostic biomarkers of cerebral cavernous angioma with symptomatic hemorrhage (CASH). Neurosurgery. 2021;88:686–97. https://doi.org/10.1093/neuros/nyaa478.
Mikati AG, Khanna O, Zhang L, Girard R, Shenkar R, Guo X, et al. Vascular permeability in cerebral cavernous malformations. J Cereb Blood Flow Metab. 2015;35:1632–9. https://doi.org/10.1038/jcbfm.2015.98.
Zeineddine HA, Girard R, Cao Y, Hobson N, Fam MD, Stadnik A, et al. Quantitative susceptibility mapping as a monitoring biomarker in cerebral cavernous malformations with recent hemorrhage. J Magn Reson Imaging. 2018;47:1133–8. https://doi.org/10.1002/jmri.25831.
Girard R, Fam MD, Zeineddine HA, Tan H, Mikati AG, Shi C, et al. Vascular permeability and iron deposition biomarkers in longitudinal follow-up of cerebral cavernous malformations. J Neurosurg. 2017;127:102–10. https://doi.org/10.3171/2016.5.JNS16687.
Huang Z, Ma L, Huang C, Li Q, Nice EC. Proteomic profiling of human plasma for cancer biomarker discovery. Proteomics. 2017;17. https://doi.org/10.1002/pmic.201600240.
Loke SY, Lee ASG. The future of blood-based biomarkers for the early detection of breast cancer. Eur J Cancer. 2018;92:54–68. https://doi.org/10.1016/j.ejca.2017.12.025.
Koskimaki J, Girard R, Li Y, Saadat L, Zeineddine HA, Lightle R, et al. Comprehensive transcriptome analysis of cerebral cavernous malformation across multiple species and genotypes. JCI Insight. 2019;4. https://doi.org/10.1172/jci.insight.126167.
Koskimaki J, Polster SP, Li Y, Romanos S, Srinath A, Zhang D, et al. Common transcriptome, plasma molecules, and imaging signatures in the aging brain and a Mendelian neurovascular disease, cerebral cavernous malformation. Geroscience. 2020;42:1351–63. https://doi.org/10.1007/s11357-020-00201-4.
Lopez-Ramirez MA, Pham A, Girard R, Wyseure T, Hale P, Yamashita A, et al. Cerebral cavernous malformations form an anticoagulant vascular domain in humans and mice. Blood. 2019;133:193–204. https://doi.org/10.1182/blood-2018-06-856062.
Lopez-Ramirez MA, Fonseca G, Zeineddine HA, Girard R, Moore T, Pham A, et al. Thrombospondin1 (TSP1) replacement prevents cerebral cavernous malformations. J Exp Med. 2017;214:3331–46. https://doi.org/10.1084/jem.20171178.
Jenny Zhou H, Qin L, Zhang H, Tang W, Ji W, He Y, et al. Endothelial exocytosis of angiopoietin-2 resulting from CCM3 deficiency contributes to cerebral cavernous malformation. Nat Med. 2016;22:1033–42. https://doi.org/10.1038/nm.4169.
Wustehube J, Bartol A, Liebler SS, Brutsch R, Zhu Y, Felbor U, et al. Cerebral cavernous malformation protein CCM1 inhibits sprouting angiogenesis by activating DELTA-NOTCH signaling. Proc Natl Acad Sci U S A. 2010;107:12640–5. https://doi.org/10.1073/pnas.1000132107.
Stockton RA, Shenkar R, Awad IA, Ginsberg MH. Cerebral cavernous malformations proteins inhibit Rho kinase to stabilize vascular integrity. J Exp Med. 2010;207:881–96. https://doi.org/10.1084/jem.20091258.
Wetzel-Strong SE, Weinsheimer S, Nelson J, Pawlikowska L, Clark D, Starr MD, et al. Pilot investigation of circulating angiogenic and inflammatory biomarkers associated with vascular malformations. Orphanet J Rare Dis. 2021;16:372. https://doi.org/10.1186/s13023-021-02009-7.
Koskimaki J, Zhang D, Li Y, Saadat L, Moore T, Lightle R, et al. Transcriptome clarifies mechanisms of lesion genesis versus progression in models of Ccm3 cerebral cavernous malformations. Acta Neuropathol Commun. 2019;7:132. https://doi.org/10.1186/s40478-019-0789-0.
Correia de Sousa M, Gjorgjieva M, Dolicka D, Sobolewski C, Foti M. Deciphering miRNAs’ action through miRNA editing. Int J Mol Sci. 2019;20. https://doi.org/10.3390/ijms20246249.
Backes C, Meese E, Keller A. Specific miRNA disease biomarkers in blood, serum and plasma: challenges and prospects. Mol Diagn Ther. 2016;20:509–18. https://doi.org/10.1007/s40291-016-0221-4.
Lugli G, Cohen AM, Bennett DA, Shah RC, Fields CJ, Hernandez AG, et al. Plasma exosomal miRNAs in persons with and without Alzheimer disease: altered expression and prospects for biomarkers. PLoS ONE. 2015;10:e0139233. https://doi.org/10.1371/journal.pone.0139233.
Kar S, Bali KK, Baisantry A, Geffers R, Samii A, Bertalanffy H. Genome-wide sequencing reveals microRNAs downregulated in cerebral cavernous malformations. J Mol Neurosci. 2017;61:178–88. https://doi.org/10.1007/s12031-017-0880-6.
Lyne SB, Girard R, Koskimaki J, Zeineddine HA, Zhang D, Cao Y, et al. Biomarkers of cavernous angioma with symptomatic hemorrhage. JCI Insight. 2019;4. https://doi.org/10.1172/jci.insight.128577.
Wendler A, Wehling M. The translatability of animal models for clinical development: biomarkers and disease models. Curr Opin Pharmacol. 2010;10:601–6. https://doi.org/10.1016/j.coph.2010.05.009.
Tang AT, Choi JP, Kotzin JJ, Yang Y, Hong CC, Hobson N, et al. Endothelial TLR4 and the microbiome drive cerebral cavernous malformations. Nature. 2017;545:305–10. https://doi.org/10.1038/nature22075.
Morrison L, Akers A. Cerebral cavernous malformation, familial. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Gripp KW, et al., editors. GeneReviews((R)). Seattle (WA)1993.
de Champfleur NM, Langlois C, Ankenbrandt WJ, Le Bars E, Leroy MA, Duffau H, et al. Magnetic resonance imaging evaluation of cerebral cavernous malformations with susceptibility-weighted imaging. Neurosurgery. 2011;68:641–7; discussion 7–8. https://doi.org/10.1227/NEU.0b013e31820773cf.
Blondal T, Brunetto MR, Cavallone D, Mikkelsen M, Thorsen M, Mang Y, et al. Genome-wide comparison of next-generation sequencing and qPCR platforms for microRNA profiling in serum. Methods Mol Biol. 2017;1580:21–44. https://doi.org/10.1007/978-1-4939-6866-4_3.
Potla P, Ali SA, Kapoor M. A bioinformatics approach to microRNA-sequencing analysis. Osteoarthritis and Cartilage Open. 2021;3:100131. https://doi.org/10.1016/j.ocarto.2020.100131.
Decker JT, Hall MS, Blaisdell RB, Schwark K, Jeruss JS, Shea LD. Dynamic microRNA activity identifies therapeutic targets in trastuzumab-resistant HER2(+) breast cancer. Biotechnol Bioeng. 2018;115:2613–23. https://doi.org/10.1002/bit.26791.
Kim SH, MacIntyre DA, Binkhamis R, Cook J, Sykes L, Bennett PR, et al. Maternal plasma miRNAs as potential biomarkers for detecting risk of small-for-gestational-age births. EBioMedicine. 2020;62:103145. https://doi.org/10.1016/j.ebiom.2020.103145.
Johnson JJ, Loeffert AC, Stokes J, Olympia RP, Bramley H, Hicks SD. Association of salivary microRNA changes with prolonged concussion symptoms. JAMA Pediatr. 2018;172:65–73. https://doi.org/10.1001/jamapediatrics.2017.3884.
Rohart F, Gautier B, Singh A, Le Cao KA. mixOmics: an R package for ‘omics feature selection and multiple data integration. PLoS Comput Biol. 2017;13:e1005752. https://doi.org/10.1371/journal.pcbi.1005752.
Dweep H, Gretz N, Sticht C. miRWalk database for miRNA-target interactions. Methods Mol Biol. 2014;1182:289–305. https://doi.org/10.1007/978-1-4939-1062-5_25.
Sticht C, De La Torre C, Parveen A, Gretz N. miRWalk: an online resource for prediction of microRNA binding sites. PLoS ONE. 2018;13:e0206239. https://doi.org/10.1371/journal.pone.0206239.
Dweep H, Sticht C, Pandey P, Gretz N. miRWalk–database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011;44:839–47. https://doi.org/10.1016/j.jbi.2011.05.002.
Sulakhe D, Balasubramanian S, Xie B, Feng B, Taylor A, Wang S, et al. Lynx: a database and knowledge extraction engine for integrative medicine. Nucleic Acids Res. 2014;42:D1007–12. https://doi.org/10.1093/nar/gkt1166.
Amur S, LaVange L, Zineh I, Buckman-Garner S, Woodcock J. Biomarker qualification: toward a multiple stakeholder framework for biomarker development, egulatory acceptance, and utilization. Clin Pharmacol Ther. 2015;98:34–46. https://doi.org/10.1002/cpt.136.
Johnston S, Gallaher Z, Czaja K. Exogenous reference gene normalization for real-time reverse transcription-polymerase chain reaction analysis under dynamic endogenous transcription. Neural Regen Res. 2012;7:1064–72. https://doi.org/10.3969/j.issn.1673-5374.2012.14.004.
Dakterzada F, Targa A, Benitez ID, Romero-ElKhayat L, de Gonzalo-Calvo D, Torres G, et al. Identification and validation of endogenous control miRNAs in plasma samples for normalization of qPCR data for Alzheimer’s disease. Alzheimers Res Ther. 2020;12:163. https://doi.org/10.1186/s13195-020-00735-x.
Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–50. https://doi.org/10.1158/0008-5472.CAN-04-0496.
Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. https://doi.org/10.1093/nar/29.9.e45.
Ruiz-Villalba A, Ruijter JM, van den Hoff MJB. Use and misuse of Cq in qPCR data analysis and reporting. Life (Basel). 2021;11. https://doi.org/10.3390/life11060496.
Selst MV, Jolicoeur P. A solution to the effect of sample size on outlier elimination. Q J Exp Psychol. 1994;47:631–50.
Davies L, Gather U. The identification of multiple outliers. J Am Stat Assoc. 1993;88:782–92.
Yuan JS, Reed A, Chen F, Stewart CN Jr. Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006;7:85. https://doi.org/10.1186/1471-2105-7-85.
Leti F, DiStefano JK. miRNA quantification method using quantitative polymerase chain reaction in conjunction with C q method. Methods Mol Biol. 2018;1706:257–65. https://doi.org/10.1007/978-1-4939-7471-9_14.
Peng F, He Ja, Loo JFC, Kong SK, Li B, Gu D. Identification of serum MicroRNAs as diagnostic biomarkers for influenza H7N9 infection. Virology Reports. 2017;7:1–8. https:/doi.org/https://doi.org/10.1016/j.virep.2016.11.001.
Dridi N, Giremus A, Giovannelli JF, Truntzer C, Hadzagic M, Charrier JP, et al. Bayesian inference for biomarker discovery in proteomics: an analytic solution. EURASIP J Bioinform Syst Biol. 2017;2017:9. https://doi.org/10.1186/s13637-017-0062-4.
Girard R, Zeineddine HA, Koskimaki J, Fam MD, Cao Y, Shi C, et al. Plasma biomarkers of inflammation and angiogenesis predict cerebral cavernous malformation symptomatic hemorrhage or lesional growth. Circ Res. 2018;122:1716–21. https://doi.org/10.1161/CIRCRESAHA.118.312680.
Girard R, Zeineddine HA, Fam MD, Mayampurath A, Cao Y, Shi C, et al. Plasma biomarkers of inflammation reflect seizures and hemorrhagic activity of cerebral cavernous malformations. Transl Stroke Res. 2018;9:34–43. https://doi.org/10.1007/s12975-017-0561-3.
Arlot S, Celisse A. A survey of cross-validation procedures for model selection. Statistics Surveys. 2010;4(40–79):40.
Xia J, Broadhurst DI, Wilson M, Wishart DS. Translational biomarker discovery in clinical metabolomics: an introductory tutorial. Metabolomics. 2013;9:280–99. https://doi.org/10.1007/s11306-012-0482-9.
Lien EC, Dibble CC, Toker A. PI3K signaling in cancer: beyond AKT. Curr Opin Cell Biol. 2017;45:62–71. https://doi.org/10.1016/j.ceb.2017.02.007.
Alzahrani AS. PI3K/Akt/mTOR inhibitors in cancer: at the bench and bedside. Semin Cancer Biol. 2019;59:125–32. https://doi.org/10.1016/j.semcancer.2019.07.009.
Chen H, Zhou L, Wu X, Li R, Wen J, Sha J, et al. The PI3K/AKT pathway in the pathogenesis of prostate cancer. Front Biosci (Landmark Ed). 2016;21:1084–91. https://doi.org/10.2741/4443.
Kar S, Samii A, Bertalanffy H. PTEN/PI3K/Akt/VEGF signaling and the cross talk to KRIT1, CCM2, and PDCD10 proteins in cerebral cavernous malformations. Neurosurg Rev. 2015;38:229–36; discussion 36–7. https://doi.org/10.1007/s10143-014-0597-8.
Martinez-Lopez A, Salvador-Rodriguez L, Montero-Vilchez T, Molina-Leyva A, Tercedor-Sanchez J, Arias-Santiago S. Vascular malformations syndromes: an update. Curr Opin Pediatr. 2019;31:747–53. https://doi.org/10.1097/MOP.0000000000000812.
Ren AA, Snellings DA, Su YS, Hong CC, Castro M, Tang AT, et al. PIK3CA and CCM mutations fuel cavernomas through a cancer-like mechanism. Nature. 2021https://doi.org/10.1038/s41586-021-03562-8
Hong T, Xiao X, Ren J, Cui B, Zong Y, Zou J, et al. Somatic MAP3K3 and PIK3CA mutations in sporadic cerebral and spinal cord cavernous malformations. Brain. 2021https://doi.org/10.1093/brain/awab117
Weng J, Yang Y, Song D, Huo R, Li H, Chen Y, et al. Somatic MAP3K3 mutation defines a subclass of cerebral cavernous malformation. Am J Hum Genet. 2021;108:942–50. https://doi.org/10.1016/j.ajhg.2021.04.005.
Lisowska J, Rodel CJ, Manet S, Miroshnikova YA, Boyault C, Planus E, et al. The CCM1-CCM2 complex controls complementary functions of ROCK1 and ROCK2 that are required for endothelial integrity. J Cell Sci. 2018;131. https://doi.org/10.1242/jcs.216093.
McKerracher L, Shenkar R, Abbinanti M, Cao Y, Peiper A, Liao JK, et al. A brain-targeted orally available ROCK2 inhibitor benefits mild and aggressive cavernous angioma disease. Transl Stroke Res. 2020;11:365–76. https://doi.org/10.1007/s12975-019-00725-8.
Wei S, Li Y, Polster SP, Weber CR, Awad IA, Shen L. Cerebral cavernous malformation proteins in barrier maintenance and regulation. Int J Mol Sci. 2020;21. https://doi.org/10.3390/ijms21020675.
Polster SP, Stadnik A, Akers AL, Cao Y, Christoforidis GA, Fam MD, et al. Atorvastatin treatment of cavernous angiomas with symptomatic hemorrhage exploratory proof of concept (AT CASH EPOC) Trial. Neurosurgery. 2019;85:843–53. https://doi.org/10.1093/neuros/nyy539.
Zhou Z, Tang AT, Wong WY, Bamezai S, Goddard LM, Shenkar R, et al. Cerebral cavernous malformations arise from endothelial gain of MEKK3-KLF2/4 signalling. Nature. 2016;532:122–6. https://doi.org/10.1038/nature17178.
Lopez-Ramirez MA, Lai CC, Soliman SI, Hale P, Pham A, Estrada EJ, et al. Astrocytes propel neurovascular dysfunction during cerebral cavernous malformation lesion formation. J Clin Invest. 2021;131. https://doi.org/10.1172/JCI139570.
Park JH, Lee JY, Shin DH, Jang KS, Kim HJ, Kong G. Loss of Mel-18 induces tumor angiogenesis through enhancing the activity and expression of HIF-1alpha mediated by the PTEN/PI3K/Akt pathway. Oncogene. 2011;30:4578–89. https://doi.org/10.1038/onc.2011.174.
Hong CC, Tang AT, Detter MR, Choi JP, Wang R, Yang X, et al. Cerebral cavernous malformations are driven by ADAMTS5 proteolysis of versican. J Exp Med. 2020;217. https://doi.org/10.1084/jem.20200140.
Liu M, Banerjee R, Rossa C Jr, D’Silva NJ. RAP1-RAC1 signaling has an important role in adhesion and migration in HNSCC. J Dent Res. 2020;99:959–68. https://doi.org/10.1177/0022034520917058.
Rho SS, Ando K, Fukuhara S. Dynamic regulation of vascular permeability by vascular endothelial cadherin-mediated endothelial cell-cell junctions. J Nippon Med Sch. 2017;84:148–59. https://doi.org/10.1272/jnms.84.148.
Li X, Zhang R, Draheim KM, Liu W, Calderwood DA, Boggon TJ. Structural basis for small G protein effector interaction of Ras-related protein 1 (Rap1) and adaptor protein Krev interaction trapped 1 (KRIT1). J Biol Chem. 2012;287:22317–27. https://doi.org/10.1074/jbc.M112.361295.
Wu M, Wang G, Tian W, Deng Y, Xu Y. MiRNA-based therapeutics for lung cancer. Curr Pharm Des. 2018;23:5989–96. https://doi.org/10.2174/1381612823666170714151715.
Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–22. https://doi.org/10.1038/nrd.2016.246.
Burns TC, Li MD, Mehta S, Awad AJ, Morgan AA. Mouse models rarely mimic the transcriptome of human neurodegenerative diseases: a systematic bioinformatics-based critique of preclinical models. Eur J Pharmacol. 2015;759:101–17. https://doi.org/10.1016/j.ejphar.2015.03.021.
Mullane K, Williams M. Preclinical models of Alzheimer’s disease: relevance and translational validity. Curr Protoc Pharmacol. 2019;84:e57. https://doi.org/10.1002/cpph.57.
Detter MR, Shenkar R, Benavides CR, Neilson CA, Moore T, Lightle R, et al. Novel murine models of cerebral cavernous malformations. Angiogenesis. 2020;23:651–66. https://doi.org/10.1007/s10456-020-09736-8.
Zeineddine HA, Girard R, Saadat L, Shen L, Lightle R, Moore T, et al. Phenotypic characterization of murine models of cerebral cavernous malformations. Lab Invest. 2019;99:319–30. https://doi.org/10.1038/s41374-018-0030-y.
Shenkar R, Venkatasubramanian PN, Wyrwicz AM, Zhao JC, Shi C, Akers A, et al. Advanced magnetic resonance imaging of cerebral cavernous malformations: part II. Imaging of lesions in murine models. Neurosurgery. 2008;63:790–7; discussion 7–8. https://doi.org/10.1227/01.NEU.0000315862.24920.49.
Nagy ZB, Bartak BK, Kalmar A, Galamb O, Wichmann B, Dank M, et al. Comparison of circulating miRNAs expression alterations in matched tissue and plasma samples during colorectal cancer progression. Pathol Oncol Res. 2019;25:97–105. https://doi.org/10.1007/s12253-017-0308-1.
Malczewska A, Kidd M, Matar S, Kos-Kudla B, Modlin IM. A comprehensive assessment of the role of miRNAs as biomarkers in gastroenteropancreatic neuroendocrine tumors. Neuroendocrinology. 2018;107:73–90. https://doi.org/10.1159/000487326.
Funding
This work was supported by grants from the NIH (R21NS087328, 5U01NS104157- 02, 1R01NS107887-01, 2 P01 NS092521-06), K01 HL133530 to MLR, William and Judith Davis Fund in Neurovascular Research to IAA, by the Be Brave for Life Foundation to RG, and the Safadi Program at the University of Chicago Translational Fellowship to RG.
Author information
Authors and Affiliations
Contributions
IAA and RG designed and conceptualized the study, oversaw data analyses, and edited the final manuscript. SR, AbS, and RG helped optimize the study design. SR, AbS, YiL, TM, JYS, RL, NH, DZ, JK, LS, SM, AgS, KP, JCP, AbSh, DS, RS, MLR, CCL, MK, DM, and MG acquired the data. SR, AbS, YiL, BX, CC, DB, YaL, DS, YJ, RG, and IAA analyzed and interpreted the data. The manuscript was written by SR, AbS, RG, IAA, and all authors edited and gave final approval of the published work.
Corresponding author
Ethics declarations
Ethics Approval
All animal experiments adhered to the NIH Guide for the Care and Use of Laboratory Animals and were approved by the respective Institutional Animal Care and Use Committees at the University of Chicago and University of California San Diego. All human studies were approved by the University of Chicago Institutional Review Board, which is guided by ethical principles consistent with the Belmont Report, and comply with the rules and regulations of the US Department of Health and Human Services Federal Policy for the Protection of Human Subjects (56 FR 28003).
Consent to Participate
All human subjects gave written informed consent in compliance with the Declaration of Helsinki.
Consent for Publication
Human research participants provided informed consent for publication of any images within the manuscript.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abhinav Srinath, Ying Li, Romuald Girard and Issam A. Awad are authors with equal contributions.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Romanos, S.G., Srinath, A., Li, Y. et al. Circulating Plasma miRNA Homologs in Mice and Humans Reflect Familial Cerebral Cavernous Malformation Disease. Transl. Stroke Res. 14, 513–529 (2023). https://doi.org/10.1007/s12975-022-01050-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12975-022-01050-3