Abstract
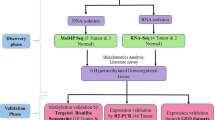
Evaluation of a cancer cell fraction is important for accurate molecular analysis, and pathological analysis is the gold standard for evaluation. Despite the potential convenience, no established molecular markers for evaluation are available. In this study, we aimed to identify ovarian cancer cell fraction markers using DNA methylation highly specific to ovarian cancer cells. Using genome-wide DNA methylation data, we screened candidate marker genes methylated in 30 ovarian cancer FFPE samples and 12 high-grade serous ovarian cancer cell lines, and unmethylated in two female leucocytes and two normal fallopian epithelial cell samples. Methylation levels of two genes, SIM1, and ZNF154, showed high correlation with pathological cancer cell fractions among the 30 ovarian cancer FFPE samples (R = 0.61 for SIM1, 0.71 for ZNF154). For cost-effective analysis of FFPE samples, pyrosequencing primers were designed, and successfully established for SIM1 and ZNF154. Correlation between a pathological cancer cell fraction and methylation levels obtained by pyrosequencing was confirmed to be high (R = 0.53 for SIM1, 0.64 for ZNF154). Finally, an independent validation cohort of 29 ovarian cancer FFPE samples was analyzed. ZNF154 methylation showed a high correlation with the pathological cancer cell fraction (R = 0.77, P < 0.0001). Therefore, the ZNF154 methylation level was considered to be useful for the estimation of ovarian cancer cell fraction, and is expected to help accurate molecular analysis.





Similar content being viewed by others
Data availability
Not available.
References
Chakravarty D, Solit DB. Clinical cancer genomic profiling. Nat Rev Genet. 2021;22(8):483–501. https://doi.org/10.1038/s41576-021-00338-8.
Ishihara H, Yamashita S, Amano R, Kimura K, Hirakawa K, Ueda T, Murakami Y, Tamori A, Tanabe K, Kawada N, Hagihara A, Ushijima T. Pancreatic cancer cell fraction estimation in a DNA sample. Oncology. 2018;95(6):370–9. https://doi.org/10.1159/000491637.
Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–15. https://doi.org/10.1038/nature10166.
Konstantinopoulos PA, Ceccaldi R, Shapiro GI, D’Andrea AD. Homologous recombination deficiency: exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov. 2015;5(11):1137–54. https://doi.org/10.1158/2159-8290.cd-15-0714.
Takahashi T, Matsuda Y, Yamashita S, Hattori N, Kushima R, Lee YC, Igaki H, Tachimori Y, Nagino M, Ushijima T. Estimation of the fraction of cancer cells in a tumor DNA sample using DNA methylation. PLoS ONE. 2013;8(12): e82302. https://doi.org/10.1371/journal.pone.0082302.
Zong L, Hattori N, Yoda Y, Yamashita S, Takeshima H, Takahashi T, Maeda M, Katai H, Nanjo S, Ando T, Seto Y, Ushijima T. Establishment of a DNA methylation marker to evaluate cancer cell fraction in gastric cancer. Gastr Cancer. 2016;19(2):361–9. https://doi.org/10.1007/s10120-015-0475-2.
Ishihara H, Yamashita S, Fujii S, Tanabe K, Mukai H, Ushijima T. DNA methylation marker to estimate the breast cancer cell fraction in DNA samples. Med Oncol. 2018;35(11):147. https://doi.org/10.1007/s12032-018-1207-3.
Salhab A, Nordström K, Gasparoni G, Kattler K, Ebert P, Ramirez F, Arrigoni L, Müller F, Polansky JK, Cadenas C, Lengauer T, Manke T, Walter J. A comprehensive analysis of 195 DNA methylomes reveals shared and cell-specific features of partially methylated domains. Genome Biol. 2018;19(1):150. https://doi.org/10.1186/s13059-018-1510-5.
Moran S, Martínez-Cardús A, Sayols S, Musulén E, Balañá C, Estival-Gonzalez A, Moutinho C, Heyn H, Diaz-Lagares A, de Moura MC, Stella GM, Comoglio PM, Ruiz-Miró M, Matias-Guiu X, Pazo-Cid R, Antón A, Lopez-Lopez R, Soler G, Longo F, Guerra I, Fernandez S, Assenov Y, Plass C, Morales R, Carles J, Bowtell D, Mileshkin L, Sia D, Tothill R, Tabernero J, Llovet JM, Esteller M. Epigenetic profiling to classify cancer of unknown primary: a multicentre, retrospective analysis. Lancet Oncol. 2016;17(10):1386–95. https://doi.org/10.1016/s1470-2045(16)30297-2.
Iorio F, Knijnenburg TA, Vis DJ, Bignell GR, Menden MP, Schubert M, Aben N, Gonçalves E, Barthorpe S, Lightfoot H, Cokelaer T, Greninger P, van Dyk E, Chang H, de Silva H, Heyn H, Deng X, Egan RK, Liu Q, Mironenko T, Mitropoulos X, Richardson L, Wang J, Zhang T, Moran S, Sayols S, Soleimani M, Tamborero D, Lopez-Bigas N, Ross-Macdonald P, Esteller M, Gray NS, Haber DA, Stratton MR, Benes CH, Wessels LFA, Saez-Rodriguez J, McDermott U, Garnett MJ. A Landscape of pharmacogenomic interactions in cancer. Cell. 2016;166(3):740–54. https://doi.org/10.1016/j.cell.2016.06.017.
Zhang W, Klinkebiel D, Barger CJ, Pandey S, Guda C, Miller A, Akers SN, Odunsi K, Karpf AR. Global DNA hypomethylation in epithelial ovarian cancer: passive demethylation and association with genomic instability. Cancers (Basel). 2020;12(3):764. https://doi.org/10.3390/cancers12030764.
Zhang Y, Long H, Wang S, Xiao W, Xiong M, Liu J, Chen L, Chen R, Wei X, Shu Y, Zeng Y, Zhang L. Genome-wide DNA methylation pattern in whole blood associated with primary intracerebral hemorrhage. Front Immunol. 2021;12: 702244. https://doi.org/10.3389/fimmu.2021.702244.
Ueda S, Yamashita S, Watanabe SI, Wakabayashi M, Motoi N, Noguchi M, Sekine S, Sato Y, Ushijima T. Influence of degree of DNA degradation in formalin-fixed and paraffin-embedded tissue samples on accuracy of genome-wide DNA methylation analysis. Epigenomics. 2021;13(8):565–76. https://doi.org/10.2217/epi-2020-0431.
Huang RL, Gu F, Kirma NB, Ruan J, Chen CL, Wang HC, Liao YP, Chang CC, Yu MH, Pilrose JM, Thompson IM, Huang HC, Huang TH, Lai HC, Nephew KP. Comprehensive methylome analysis of ovarian tumors reveals hedgehog signaling pathway regulators as prognostic DNA methylation biomarkers. Epigenetics. 2013;8(6):624–34. https://doi.org/10.4161/epi.24816.
Kamimae S, Yamamoto E, Kai M, Niinuma T, Yamano HO, Nojima M, Yoshikawa K, Kimura T, Takagi R, Harada E, Harada T, Maruyama R, Sasaki Y, Tokino T, Shinomura Y, Sugai T, Imai K, Suzuki H. Epigenetic silencing of NTSR1 is associated with lateral and noninvasive growth of colorectal tumors. Oncotarget. 2015;6(30):29975–90. https://doi.org/10.18632/oncotarget.5034.
Xu Z, Gujar H, Fu G, Ahmadi H, Bhanvadia S, Weisenberger DJ, Jin B, Gill PS, Gill I, Daneshmand S, Siegmund KD, Liang G. A novel DNA methylation signature as an independent prognostic factor in muscle-invasive bladder cancer. Front Oncol. 2021;11: 614927. https://doi.org/10.3389/fonc.2021.614927.
Sánchez-Vega F, Gotea V, Petrykowska HM, Margolin G, Krivak TC, DeLoia JA, Bell DW, Elnitski L. Recurrent patterns of DNA methylation in the ZNF154, CASP8, and VHL promoters across a wide spectrum of human solid epithelial tumors and cancer cell lines. Epigenetics. 2013;8(12):1355–72. https://doi.org/10.4161/epi.26701.
Margolin G, Petrykowska HM, Jameel N, Bell DW, Young AC, Elnitski L. Robust detection of DNA hypermethylation of ZNF154 as a pan-cancer locus with in silico modeling for blood-based diagnostic development. J Mol Diagn. 2016;18(2):283–98. https://doi.org/10.1016/j.jmoldx.2015.11.004.
Miller BF, Petrykowska HM, Elnitski L. Assessing ZNF154 methylation in patient plasma as a multicancer marker in liquid biopsies from colon, liver, ovarian and pancreatic cancer patients. Sci Rep. 2021;11(1):221. https://doi.org/10.1038/s41598-020-80345-7.
Ogasawara A, Hihara T, Shintani D, Yabuno A, Ikeda Y, Tai K, Fujiwara K, Watanabe K, Hasegawa K. Evaluation of circulating tumor DNA in patients with ovarian cancer harboring somatic PIK3CA or KRAS mutations. Cancer Res Treat. 2020;52(4):1219–28. https://doi.org/10.4143/crt.2019.688.
Kim S, Han Y, Kim SI, Kim HS, Kim SJ, Song YS. Tumor evolution and chemoresistance in ovarian cancer. NPJ Precis Oncol. 2018;2:20. https://doi.org/10.1038/s41698-018-0063-0.
Acknowledgements
The authors are grateful to Dr. K. Ichimura, Ms. Y. Matsushita, and Ms. M. Kitahara of Division of Brain Tumor Translational Research in National Cancer Center Research Institute for their technical assistance with the experiments. This study was supported by AMED JP21cm0106451, JP21ck0106466 and JSPS KAKENHI JP20K18180.
Funding
This study was supported by AMED JP21cm0106451, JP21ck0106466 and JSPS KAKENHI JP20K18180.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Data collection and analysis were performed by TE, SY and HY. The first draft of the manuscript was written by TE. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
The authors state no conflicts of interest regarding this work.
Ethical approval
This study protocol was approved by the institutional review board at each participating center. This study was performed in accordance with the ethical standards of 1964 Declaration of Helsinki and its later amendments.
Informed consent
Written informed consent was obtained from all participants.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
12032_2022_1679_MOESM1_ESM.pptx
Supplementary file1 (PPTX 63 kb). Supplementary Figure S1. Methylation levels of candidate marker genes in clinicalsamples.Methylation levels of SIM1, OTX2, and ZNF154 obtained by DNA methylationmicroarray analysis are shown for female leucocyte (n = 2), normal fallopian epidermalcell (n = 2) and ovarian cancer samples (n = 30). These three genes were considered to bemethylated in cancer samples and unmethylated in normal samples, and such a profilewas observed overall. However, OTX2 tended to show lower methylation levels than theother two genes, and was excluded from further analysis.Supplementary Figure 2. Correlation between methylation levels of candidatemarker genes obtained by DNA methylation array and pyrosequencing.Samples in the screening cohort with the remaining were analyzed bypyrosequencing, and correlation between the methylation levels of (A) SIM1 and (B) ZNF154 obtained by pyrosequencing and those by DNA methylation microarray wasanalyzed. While ZNF154 showed a high correlation, SIM1 showed only a moderatecorrelation.
Rights and permissions
About this article
Cite this article
Ebata, T., Yamashita, S., Takeshima, H. et al. DNA methylation marker to estimate ovarian cancer cell fraction. Med Oncol 39, 78 (2022). https://doi.org/10.1007/s12032-022-01679-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-022-01679-y