Abstract
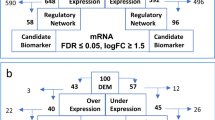
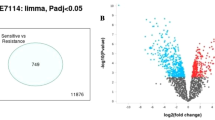
Multiple myeloma (MM), second most common hematological malignancy, still remains irremediable because of acquisition of drug resistance. Glucocorticoid (GC) therapy, which is used as one of the key therapies against MM, is hindered by the incidence of GC resistance. The underlying mechanism of this acquired GC resistance in MM is not fully elucidated. Therefore, the present study was aimed to identify the differentially expressed genes (DEGs), associated micro RNAs (miRNAs), and transcription factors (TFs) from the microarray datasets of GC-resistant and GC-sensitive MM cell lines, obtained from Gene Expression Omnibus (GEO) database. DEGs were identified using GEO2R tool from two datasets and common DEGs were obtained by constructing Venn diagram. Then the Gene ontology (GO) and pathway enrichment analysis were performed using DAVID database. Genetic alterations in DEGs were examined using COSMIC database. Protein–protein interaction (PPI) network of DEGs was constructed using STRING database and Cytoscape tool. Network of interaction of DEGs and miRNAs as well as TFs were obtained and constructed using mirDIP, TRRUST, and miRNet tools. Drug gene interaction was studied to identify potential drug molecules by DGIdb tool. Six common DEGs, CDKN1A, CDKN2A, NLRP11, BTK, CD52, and RELN, were found to be significantly upregulated in GC-resistant MM and selected for further analysis. miRNA analysis detected hsa-mir-34a-5p that could interact with maximum target DEGs. Two TFs, Sp1 and Sp3, were found to regulate the expression of selected DEGs. The entire study may provide a new understanding about the GC resistance in MM.






Similar content being viewed by others
Availability of data and materials
The datasets analyzed in the present study are available in the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/).
Code availability
Not applicable.
References
Rajkumar SV, Dimopoulos MA, Palumbo A, et al. International Myeloma Working Group Updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 2014;15:e538–48. https://doi.org/10.1002/ajh.25791.
Palumbo A, Anderson K. Multiple myeloma. N Engl J Med. 2011;364(11):1046–60. https://doi.org/10.1056/NEJMra1011442.
Kyle RA, Rajkumar SV. Multiple myeloma. N Engl J Med. 2004;351:1860–73. https://doi.org/10.1056/NEJMra041875.s.
Kyle RA, Therneau TM, Rajkumar SV, et al. Prevalence of monoclonal gammopathy of undetermined significance. N Engl J Med. 2006;354(13):1362–9. https://doi.org/10.1056/NEJMoa054494.
Landgren O, Kyle RA, Pfeiffer RM, et al. Monoclonal gammopathy of undetermined significance (MGUS) consistently precedes multiple myeloma: a prospective study. Blood. 2009;113(22):5412–7. https://doi.org/10.1182/blood-2008-12-194241.
Iida S. Mechanisms of action and resistance for multiple myeloma novel drug treatments. Int J Hematol. 2016;104(3):271–2. https://doi.org/10.1007/s12185-016-2040-0.
Chauhan D, Pandey P, Ogata A, et al. Dexamethasone induces apoptosis of multiple myeloma cells in a JNK/SAP kinase independent mechanism. Oncogene. 1997;15(7):837–43. https://doi.org/10.1038/sj.onc.1201253.
Burwick N, Sharma S. Glucocorticoids in multiple myeloma: past, present, and future. Ann Hematol. 2019;98(1):19–28. https://doi.org/10.1007/s00277-018-3465-8.
Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–10. https://doi.org/10.1093/nar/30.1.207.
Palagani A, Op de Beeck K, Naulaerts S, et al. Ectopic microRNA-150-5p transcription sensitizes glucocorticoid therapy response in MM1S multiple myeloma cells but fails to overcome hormone therapy resistance in MM1R cells. PLoS ONE. 2014;9(12):e113842. https://doi.org/10.1371/journal.pone.0113842.
Thomas AL, Coarfa C, Qian J, et al. Identification of potential glucocorticoid receptor therapeutic targets in multiple myeloma. Nucl Recept Signal. 2015;13:e006. https://doi.org/10.1621/nrs.13006.
Logie E, Chirumamilla CS, Perez-Novo C, et al. Covalent cysteine targeting of Bruton’s Tyrosine Kinase (BTK) family by Withaferin-A reduces survival of glucocorticoid-resistant multiple myeloma MM1 cells. Cancers (Basel). 2021;13(7):1618. https://doi.org/10.3390/cancers13071618.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research. 2015;43:e47e47. https://doi.org/10.1093/nar/gkv007.
Da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. https://doi.org/10.1038/nprot.2008.211.
Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2010;39:D945–50. https://doi.org/10.1093/nar/gkq929.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:p1. https://doi.org/10.1126/scisignal.6273er1.
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–13. https://doi.org/10.1093/nar/gky1131.
Almeida D, Azevedo V, Silva A, Baumbach J. PetriScape—a plugin for discrete Petri net simulations in Cytoscape. J Integr Bioinform. 2016;13(1):284. https://doi.org/10.2390/biecoll-jib-2016-284.
Tokar T, Pastrello C, Rossos AEM, Abovsky M, Hauschild AC, Tsay M, Lu R, Jurisica I. mirDIP 41-integrative database of human microRNA target predictions. Nucleic Acids Res. 2018;46(D1):D360–70. https://doi.org/10.1093/nar/gkx1144.
Han H, Cho JW, Lee S, Yun A, Kim H, Bae D, Yang S, Kim CY, Lee M, Kim E, Lee S, Kang B, Jeong D, Kim Y, Jeon HN, Jung H, Nam S, Chung M, Kim JH, Lee I. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018;46:D380–6. https://doi.org/10.1093/nar/gkx1013.
Fan Y, Siklenka K, Arora SK, Ribeiro P, Kimmins S, Xia J. miRNet - dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res. 2016;44(W1):W135–41. https://doi.org/10.1093/nar/gkw288.
Cotto KC, Wagner AH, Feng Y-Y, Kiwala S, Coffman AC, Spies G, Wollam A, Spies NC, Griffith OL, Griffith M. DGIdb 3.0: a redesign and expansion of the drug-gene interaction database. Nucleic Acids Res. 2018;46:D1068–73. https://doi.org/10.1093/nar/gkx1143.
Wu D, Wang X. Application of clinical bioinformatics in lung cancer-specific biomarkers. Cancer Metastasis Rev. 2015;34(2):209–16. https://doi.org/10.1007/s10555-015-9564-2.
Mohr S, Leikauf GD, Keith G, Rihn BH. Microarrays as cancer keys: an array of possibilities. J Clin Oncol. 2002;20:31653175. https://doi.org/10.1200/jco.2002.12.073.
Hideshima T, Anderson KC. Signaling pathway mediating myeloma cell growth and survival. Cancers (Basel). 2021;13(2):216. https://doi.org/10.3390/cancers1302021.
Jovanović KK, Escure G, Demonchy J, et al. Deregulation and targeting of TP53 pathway in multiple myeloma. Front Oncol. 2019;8:665. https://doi.org/10.3389/fonc.2018.00665.
van Andel H, Kocemba KA, Spaargaren M, Pals ST. Aberrant Wnt signaling in multiple myeloma: molecular mechanisms and targeting options. Leukemia. 2019;33(5):1063–75. https://doi.org/10.1038/s41375-019-0404-1.
Stelzl U, Worm U, Lalowski M, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122(6):957–68. https://doi.org/10.1016/j.cell.2005.08.029.
Kreis NN, Louwen F, Yuan J. The multifaceted p21 (Cip1/Waf1/CDKN1A) in cell differentiation, migration and cancer therapy. Cancers (Basel). 2019;11(9):1220. https://doi.org/10.3390/cancers11091220.
Huang Y, Wang W, Chen Y, et al. The opposite prognostic significance of nuclear and cytoplasmic p21 expression in resectable gastric cancer patients. J Gastroenterol. 2014;49(11):1441–52. https://doi.org/10.1007/s00535-013-0900-4.
Liu R, Wettersten HI, Park SH, Weiss RH. Small-molecule inhibitors of p21 as novel therapeutics for chemotherapy-resistant kidney cancer. Future Med Chem. 2013;5(9):991–4. https://doi.org/10.4155/fmc.13.56.
Romagosa C, Simonetti S, López-Vicente L, et al. p16(Ink4a) overexpression in cancer: a tumor suppressor gene associated with senescence and high-grade tumors. Oncogene. 2011;30(18):2087–97. https://doi.org/10.1038/onc.2010.614.
Coordes A, Lenz K, Qian X, Lenarz M, Kaufmann AM, Albers AE. Meta-analysis of survival in patients with HNSCC discriminates risk depending on combined HPV and p16 status. Eur Arch Otorhinolaryngol. 2016;273(8):2157–69. https://doi.org/10.1007/s00405-015-3728-0.
Ellwanger K, Becker E, Kienes I, et al. The NLR family pyrin domain-containing 11 protein contributes to the regulation of inflammatory signaling. J Biol Chem. 2018;293(8):2701–10. https://doi.org/10.1074/jbc.RA117.000152.
Von Suskil M, Sultana KN, Elbezanti WO, et al. Bruton’s tyrosine kinase targeting in multiple myeloma. Int J Mol Sci. 2021;22(11):5707. https://doi.org/10.3390/ijms22115707.
Yang Y, Shi J, Gu Z, et al. Bruton tyrosine kinase is a therapeutic target in stem-like cells from multiple myeloma. Cancer Res. 2015;75(3):594–604. https://doi.org/10.1158/0008-5472.CAN-14-2362.
Richardson PG, Bensinger WI, Huff CA, et al. Ibrutinib alone or with dexamethasone for relapsed or relapsed and refractory multiple myeloma: phase 2 trial results. Br J Haematol. 2018;180(6):821–30. https://doi.org/10.1111/bjh.15058.
Saito Y, Nakahata S, Yamakawa N, et al. CD52 as a molecular target for immunotherapy to treat acute myeloid leukemia with high EVI1 expression. Leukemia. 2011;25(6):921–31. https://doi.org/10.1038/leu.2011.36.
Wang J, Zhang G, Sui Y, et al. CD52 Is a Prognostic Biomarker and Associated With Tumor Microenvironment in Breast Cancer. Front Genet. 2020;11:578002. https://doi.org/10.3389/fgene.2020.578002.
Lin L, Yan F, Zhao D, et al. Reelin promotes the adhesion and drug resistance of multiple myeloma cells via integrin β1 signaling and STAT3. Oncotarget. 2016;7(9):9844–58. https://doi.org/10.18632/oncotarget.7151.
Qin X, Lin L, Cao L, et al. Extracellular matrix protein Reelin promotes myeloma progression by facilitating tumor cell proliferation and glycolysis. Sci Rep. 2017;7:45305. https://doi.org/10.1038/srep45305.
Lin L, Wang P, Liu X, et al. Epigenetic regulation of reelin expression in multiple myeloma. Hematol Oncol. 2017;35(4):685–92. https://doi.org/10.1002/hon.2311.
Dou A, Zhang Y, Wang Y, Liu X, Guo Y. Reelin depletion alleviates multiple myeloma bone disease by promoting osteogenesis and inhibiting osteolysis. Cell Death Discov. 2021;7(1):219. https://doi.org/10.1038/s41420-021-00608-8.
Zhang L, Liao Y, Tang L. MicroRNA-34 family: a potential tumor suppressor and therapeutic candidate in cancer. J Exp Clin Cancer Res. 2019;38(1):53. https://doi.org/10.1186/s13046-019-1059-5.
Xiao X, Gu Y, Wang G, Chen S. c-Myc, RMRP, and miR-34a-5p form a positive-feedback loop to regulate cell proliferation and apoptosis in multiple myeloma. Int J Biol Macromol. 2019;122:526–37. https://doi.org/10.1016/j.ijbiomac.2018.10.207.
Jin Z, Zhou S, Ye H, Jiang S, Yu K, Ma Y. The mechanism of SP1/p300 complex promotes proliferation of multiple myeloma cells through regulating IQGAP1 transcription. Biomed Pharmacother. 2019;119:109434. https://doi.org/10.1016/j.biopha.2019.109434.
Karki K, Harishchandra S, Safe S. Bortezomib Targets Sp Transcription Factors in Cancer Cells. Mol Pharmacol. 2018;94(4):1187–96. https://doi.org/10.1124/mol.118.112797.
Wang B, Lyu H, Pei S, Song D, Ni J, Liu B. Cladribine in combination with entinostat synergistically elicits anti-proliferative/anti-survival effects on multiple myeloma cells. Cell Cycle. 2018;17(8):985–96. https://doi.org/10.1080/15384101.2018.1464849.
Funding
No funding was received to conduct this study.
Author information
Authors and Affiliations
Contributions
SG conceptualized the study, performed the analysis, and wrote the manuscript. SB supervised the study.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ghosal, S., Banerjee, S. In silico bioinformatics analysis for identification of differentially expressed genes and therapeutic drug molecules in Glucocorticoid-resistant Multiple myeloma. Med Oncol 39, 53 (2022). https://doi.org/10.1007/s12032-022-01651-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-022-01651-w