Abstract
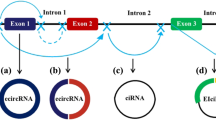
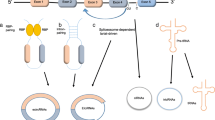
Circular RNAs (circRNAs) are a unique family of endogenous RNAs devoid of 3′ poly-A tails and 5′ end caps. These single-stranded circRNAs, found in the cytoplasm, are synthesized via back-splicing mechanisms, merging introns, exons, or both, resulting in covalently closed circular loops. They are profusely expressed across the eukaryotic transcriptome and offer heightened stability against exonuclease RNase R compared to linear RNA counterparts. This review endeavors to provide a comprehensive overview of circRNAs’ characteristics, biogenesis, and mechanisms of action. Furthermore, aimed to shed light on the potential of circRNAs as significant biomarkers in various cancer types. It has been performed an exhaustive literature review, drawing on recent studies and findings related to circRNA characteristics, synthesis, function, evaluation techniques, and their associations with oncogenesis. CircRNAs are intricately associated with tumor progression and development. Their multifaceted roles encompass gene regulation through the sponging of proteins and microRNAs, controlling transcription and splicing, interacting with RNA binding proteins (RBPs), and facilitating gene translation. Due to these varied roles, circRNAs have become a focal point in tumor pathology investigations, given their promising potential as both biomarkers and therapeutic agents. CircRNAs, due to their unique biogenesis and multifunctionality, hold immense promise in the realm of oncology. Their stability, widespread expression, and intricate involvement in gene regulation underscore their prospective utility as reliable biomarkers and therapeutic targets in cancer. As our understanding of circRNAs deepens, advanced techniques for their detection, evaluation, and manipulation will likely emerge. These advancements might catalyze the translation of circRNA-based diagnostics and therapeutics into clinical practice, potentially revolutionizing cancer care and prognosis.


Similar content being viewed by others
Data availability
Not applicable.
Abbreviations
- ALS:
-
Amyotrophic lateral sclerosis
- ANKRD52:
-
Ankyrin repeat domain 52
- BWA-MEM:
-
Burrow-wheeler aligner’s maximal exact match
- CDR1as:
-
Cerebellar degeneration-related protein 1 antisense
- ceRNA:
-
Competitive endogenous RNA
- CDK2:
-
Cyclin-dependent kinase 2
- circRNA:
-
Circular RNA
- circ-Foxo3:
-
Circular RNA forkhead box O3
- circHIPK3:
-
Circular RNA homeodomain-interacting protein kinase 3
- circ-MBL:
-
Circular muscleblind
- circMTO1:
-
Circular RNA mitochondrial tRNA translation optimization 1
- circ-ZNF609:
-
Circular RNA zinc finger protein 609
- CIRI:
-
Circular RNA identifier
- CSCD:
-
Cancer-specific circRNA database
- EIciRNAs:
-
Exon–intron circRNAs
- IRES:
-
Internal ribosome entry site
- KNIFE:
-
Known and novel isoform explorer
- MREs:
-
MiRNA response elements
- miRNA:
-
MicroRNA
- ncRNAs:
-
Non-coding RNA
- NCLScan:
-
Non-co-linear transcripts scan
- PTESFinder:
-
Post-transcriptional exon shuffling finder
- RBPs:
-
RNA binding proteins
- RPAD:
-
RNase R protocol accompanied by poly (A) + RNA depletion and polyadenylation
- TDP-43:
-
TAR DNA-binding protein 43
- TSCD:
-
Tissue-specific CircRNA database
References
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK et al (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495(7441):384–388
Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK (1976) Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci 73(11):3852–3856
Wu W, Ji P, Zhao F (2020) CircAtlas: an integrated resource of one million highly accurate circular RNAs from 1070 vertebrate transcriptomes. Genome Biol 21(1):1–14
Kristensen L, Andersen M, Stagsted L, Ebbesen K, Hansen T, Kjems J (2019) The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 20(11):675–691
Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO (2012) Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 7(2):e30733
Barrett SP, Salzman J (2016) Circular RNAs: analysis, expression and potential functions. Development 143(11):1838
Han D, Li J, Wang H, Su X, Hou J, Gu Y et al (2017) Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology 66(4):1151–1164
Chen LL, Yang L (2015) Regulation of circRNA biogenesis. RNA Biol 12(4):381–388
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A et al (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495(7441):333–338
Xiao J (2018) Circular RNAs: biogenesis and functions. Springer, Singapore
Wilusz J, Sharp PJS (2013) A circuitous route to noncoding RNA NIH public access. Science 340:440–441
Huang A, Zheng H, Wu Z, Chen M, Huang Y (2020) Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics 10(8):3503
Liu J, Zhang X, Yan M, Li H (2020) Emerging role of circular RNAs in cancer. Front Oncol 10:663
Huang R, Zhang Y, Han B, Bai Y, Zhou R, Gan G et al (2017) Circular RNA HIPK2 regulates astrocyte activation via cooperation of autophagy and ER stress by targeting MIR124–2HG. Autophagy 13(10):1722–1741
Bach DH, Lee SK, Sood AK (2019) Circular RNAs in cancer. Mol Therap Nucleic Acids 16:118–129
Guo JU, Agarwal V, Guo H, Bartel DP (2014) Expanded identification and characterization of mammalian circular RNAs. Genom Biol 15(7):1–14
Zhu J, Yu Y, Meng X, Fan Y, Zhang Y, Zhou C et al (2013) De novo-generated small palindromes are characteristic of amplicon boundary junction of double minutes. Int J Cancer 133(4):797–806
Lasda E, Parker R (2014) Circular RNAs: diversity of form and function. RNA 20(12):1829–1842
Hayes M, Li J (2015) An integrative framework for the identification of double minute chromosomes using next generation sequencing data. BMC Genet 16(2):1–7
Tan S, Gou Q, Pu W, Guo C, Yang Y, Wu K et al (2018) Circular RNA F-circEA produced from EML4-ALK fusion gene as a novel liquid biopsy biomarker for non-small cell lung cancer. Cell Res 28(6):693–695
Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO (2013) Cell-type specific features of circular RNA expression. PLoS Genet 9(9):e1003777
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB (2016) Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 44(6):2846–2858
Preußer C, Hung LH, Schneider T, Schreiner S, Hardt M, Moebus A et al (2018) Selective release of circRNAs in platelet-derived extracellular vesicles. J Extracell Vesicles 7(1):1424473
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M et al (2014) circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56(1):55–66
Zhu LP, He YJ, Hou JC, Chen X, Zhou SY, Yang SJ et al (2017) The role of circRNAs in cancers. Biosci Rep. https://doi.org/10.1042/BSR20170750
Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P et al (2014) Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep 9(5):1966–1980
Chen LL (2016) The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 17(4):205–211
Shen T, Han M, Wei G, Ni T (2015) An intriguing RNA species—perspectives of circularized RNA. Protein Cell 6(12):871–880
Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J et al (2016) Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res 26(9):1277–1287
Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA et al (2015) The RNA binding protein quaking regulates formation of circRNAs. Cell 160(6):1125–1134
Tang X, Ren H, Guo M, Qian J, Yang Y, Gu C et al (2021) Review on circular RNAs and new insights into their roles in cancer. Cell 19:910–928
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L (2014) Complementary sequence-mediated exon circularization. Cell 159(1):134–147
Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR et al (2015) Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 10(2):170–177
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH et al (2013) Circular intronic long noncoding RNAs. Mol Cell 51(6):792–806
Kelly S, Greenman C, Cook PR, Papantonis A (2015) Exon skipping is correlated with exon circularization. J Mol Biol 427(15):2414–2417
Ebert MS, Sharp PA (2010) MicroRNA sponges: progress and possibilities. RNA 16(11):2043–2050
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP (2011) A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146(3):353–358
Piwecka M, Glažar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A et al (2017) Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357(6357):8526
Kohlhapp FJ, Mitra AK, Lengyel E, Peter ME (2015) MicroRNAs as mediators and communicators between cancer cells and the tumor microenvironment. Oncogene 34(48):5857–5868
Zhao J, He S, Minassian A, Li J, Feng P (2015) Recent advances on viral manipulation of NF-κB signaling pathway. Curr Opin Virol 15:103–111
Hansen TB, Kjems J, Damgaard CK (2013) Circular RNA and miR-7 in CancerCircular RNA and miR-7 in Cancer. Cancer Res 73(18):5609–5612
Militello G, Weirick T, John D, Döring C, Dimmeler S, Uchida S (2017) Screening and validation of lncRNAs and circRNAs as miRNA sponges. Brief Bioinfo 18(5):780–788
Ikeda Y, Morikawa S, Nakashima M, Yoshikawa S, Taniguchi K, Sawamura H et al (2023) CircRNAs and RNA-binding proteins involved in the pathogenesis of cancers or central nervous system disorders. Noncoding RNA. https://doi.org/10.3390/ncrna9020023
Zhao X, Zhong Y, Wang X, Shen J, An W (2022) Advances in circular RNA and its applications. Int J Med Sci 19(6):975–985. https://doi.org/10.7150/ijms.71840
Zang J, Lu D, Xu A (2020) The interaction of circRNAs and RNA binding proteins: an important part of circRNA maintenance and function. J Neurosci Res 98(1):87–97. https://doi.org/10.1002/jnr.24356
Wawrzyniak O, Zarębska Ż, Kuczyński K, Gotz-Więckowska A, Rolle K (2020) Protein-related circular RNAs in human pathologies. Cells. https://doi.org/10.3390/cells9081841
Armakola M, Higgins MJ, Figley MD, Barmada SJ, Scarborough EA, Diaz Z et al (2012) Inhibition of RNA lariat debranching enzyme suppresses TDP-43 toxicity in ALS disease models. Nat Genet 44(12):1302–1309
Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O et al (2017) Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell 66(1):22–37
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y et al (2017) Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res 27(5):626–641
Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L et al (2017) Translation of circRNAs. Mol Cell 66(1):9–21
Chen L, Shan G (2021) CircRNA in cancer: Fundamental mechanism and clinical potential. Cancer Lett 505:49–57
Xiao W, Li J, Hu J, Wang L, Huang JR, Sethi G et al (2021) Circular RNAs in cell cycle regulation: Mechanisms to clinical significance. Cell Prolif 54(12):e13143. https://doi.org/10.1111/cpr.13143
Huang Y, Zhang C, Xiong J, Ren H (2021) Emerging important roles of circRNAs in human cancer and other diseases. Genes Dis 8(4):412–423. https://doi.org/10.1016/j.gendis.2020.07.012
Zhang Y, Luo J, Yang W, Ye WC (2023) CircRNAs in colorectal cancer: potential biomarkers and therapeutic targets. Cell Death Dis 14(6):353. https://doi.org/10.1038/s41419-023-05881-2
Zhang F, Li L, Fan Z (2022) circRNAs and their relationship with breast cancer: a review. World J Surg Oncol 20(1):373. https://doi.org/10.1186/s12957-022-02842-5
Zhang N, Nan A, Chen L, Li X, Jia Y, Qiu M et al (2020) Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Mol Cancer 19(1):101. https://doi.org/10.1186/s12943-020-01221-6
Zhou Y, Mao X, Peng R, Bai D (2022) CircRNAs in hepatocellular carcinoma: characteristic, functions and clinical significance. Int J Med Sci 19(14):2033–2043. https://doi.org/10.7150/ijms.74713
Tang K, Zhang H, Li Y, Sun Q, Jin H (2021) Circular RNA as a potential biomarker for melanoma: a systematic review. Front Cell Dev Biol 9:638548. https://doi.org/10.3389/fcell.2021.638548
Ng WL, Mohd Mohidin TB, Shukla K (2018) Functional role of circular RNAs in cancer development and progression. RNA Biol 15(8):995–1005. https://doi.org/10.1080/15476286.2018.1486659
Riquelme I, Tapia O, Espinoza JA, Leal P, Buchegger K, Sandoval A et al (2016) The gene expression status of the PI3K/AKT/mTOR pathway in gastric cancer tissues and cell lines. Pathol Oncol Res 22(4):797–805. https://doi.org/10.1007/s12253-016-0066-5
Dou Z, Gao L, Ren W, Zhang H, Wang X, Li S et al (2020) CiRS-7 functions as a ceRNA of RAF-1/PIK3CD to promote metastatic progression of oral squamous cell carcinoma via MAPK/AKT signaling pathways. Exp Cell Res 396(2):112290. https://doi.org/10.1016/j.yexcr.2020.112290
Yang X, Li S, Wu Y, Ge F, Chen Y, Xiong Q (2020) The circular RNA CDR1as regulate cell proliferation via TMED2 and TMED10. BMC Cancer 20(1):312. https://doi.org/10.1186/s12885-020-06794-5
Jiang M, Fang S, Zhao X, Zhou C, Gong Z (2021) Epithelial-mesenchymal transition-related circular RNAs in lung carcinoma. Cancer Biol Med 18(2):411–420. https://doi.org/10.20892/j.issn.2095-3941.2020.0238
Wu Z, Yu X, Zhang S, He Y, Guo W (2022) Mechanism underlying circRNA dysregulation in the TME of digestive system cancer. Front Immunol. https://doi.org/10.3389/fimmu.2022.951561
Shao Y, Lu B (2020) The crosstalk between circular RNAs and the tumor microenvironment in cancer metastasis. Cancer Cell Int 20:448. https://doi.org/10.1186/s12935-020-01532-0
Begliarzade S, Sufianov A, Ilyasova T, Shumadalova A, Sufianov R, Beylerli O et al (2024) Circular RNA in cervical cancer: fundamental mechanism and clinical potential. Noncoding RNA Res 9(1):116–124. https://doi.org/10.1016/j.ncrna.2023.11.009
He AT, Liu J, Li F, Yang BB (2021) Targeting circular RNAs as a therapeutic approach: current strategies and challenges. Signal Transduct Target Ther 6(1):185. https://doi.org/10.1038/s41392-021-00569-5
Zhang F, Jiang J, Qian H, Yan Y, Xu W (2023) Exosomal circRNA: emerging insights into cancer progression and clinical application potential. J Hematol Oncol 16(1):67. https://doi.org/10.1186/s13045-023-01452-2
Dhuri K, Bechtold C, Quijano E, Pham H, Gupta A, Vikram A et al (2020) Antisense oligonucleotides: an emerging area in drug discovery and development. J Clin Med. https://doi.org/10.3390/jcm9062004
Zheng XB, Zhang M, Xu MQ (2017) Detection and characterization of ciRS-7: a potential promoter of the development of cancer. Neoplasma 64(3):321–328. https://doi.org/10.4149/neo_2017_301
Sharma AR, Bhattacharya M, Bhakta S, Saha A, Lee SS, Chakraborty C (2021) Recent research progress on circular RNAs: biogenesis, properties, functions, and therapeutic potential. Mol Ther Nucleic Acids 25:355–371. https://doi.org/10.1016/j.omtn.2021.05.022
Pisignano G, Michael DC, Visal TH, Pirlog R, Ladomery M, Calin GA (2023) Going circular: history, present, and future of circRNAs in cancer. Oncogene 42(38):2783–2800. https://doi.org/10.1038/s41388-023-02780-w
Jagtap U, Anderson ES, Slack FJ (2023) The emerging value of circular noncoding RNA research in cancer diagnosis and treatment. Cancer Res 83(6):809–813. https://doi.org/10.1158/0008-5472.Can-22-3014
Memczak S, Papavasileiou P, Peters O, Rajewsky N (2015) Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS ONE 10(10):e0141214
Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A (2006) Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res 34(8):e63
Qu S, Liu Z, Yang X, Zhou J, Yu H, Zhang R et al (2018) The emerging functions and roles of circular RNAs in cancer. Cancer Lett 414:301–309
Li J, Li Z, Jiang P, Peng M, Zhang X, Chen K et al (2018) Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res 37(1):1–16
Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y et al (2015) The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem 61(1):221–230
Wang R, Zhang S, Chen X, Li N, Li J, Jia R et al (2018) CircNT5E acts as a sponge of miR-422a to promote glioblastoma tumorigenesis. Cancer Res 78(17):4812–4825. https://doi.org/10.1158/0008-5472.Can-18-0532
Wang R, Zhang S, Chen X, Li N, Li J, Jia R et al (2018) EIF4A3-induced circular RNA MMP9 (circMMP9) acts as a sponge of miR-124 and promotes glioblastoma multiforme cell tumorigenesis. Mol Cancer 17(1):166. https://doi.org/10.1186/s12943-018-0911-0
Lei Y, Chen L, Zhang G, Shan A, Ye C, Liang B et al (2020) MicroRNAs target the Wnt/β-catenin signaling pathway to regulate epithelial-mesenchymal transition in cancer (Review). Oncol Rep 44(4):1299–1313. https://doi.org/10.3892/or.2020.7703
Liu W, Ma W, Yuan Y, Zhang Y, Sun S (2018) Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. Biochem Biophys Res Commun 500(4):846–851. https://doi.org/10.1016/j.bbrc.2018.04.172
Chen L, Nan A, Zhang N, Jia Y, Li X, Ling Y et al (2019) Circular RNA 100146 functions as an oncogene through direct binding to miR-361-3p and miR-615-5p in non-small cell lung cancer. Mol Cancer 18(1):13. https://doi.org/10.1186/s12943-019-0943-0
Tang H, Huang X, Wang J, Yang L, Kong Y, Gao G et al (2019) circKIF4A acts as a prognostic factor and mediator to regulate the progression of triple-negative breast cancer. Mol Cancer 18(1):23. https://doi.org/10.1186/s12943-019-0946-x
Zhang X, Wang S, Wang H, Cao J, Huang X, Chen Z et al (2019) Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer 18(1):20. https://doi.org/10.1186/s12943-018-0935-5
Li RC, Ke S, Meng FK, Lu J, Zou XJ, He ZG et al (2018) CiRS-7 promotes growth and metastasis of esophageal squamous cell carcinoma via regulation of miR-7/HOXB13. Cell Death Dis 9(8):838. https://doi.org/10.1038/s41419-018-0852-y
Xie F, Li Y, Wang M, Huang C, Tao D, Zheng F et al (2018) Circular RNA BCRC-3 suppresses bladder cancer proliferation through miR-182-5p/p27 axis. Mol Cancer 17(1):144. https://doi.org/10.1186/s12943-018-0892-z
Rao D, Yu C, Sheng J, Lv E, Huang W (2021) The emerging roles of circFOXO3 in cancer. Front Cell Dev Biol 9:659417. https://doi.org/10.3389/fcell.2021.659417
Zhang L, Zhou Q, Qiu Q, Hou L, Wu M, Li J et al (2019) CircPLEKHM3 acts as a tumor suppressor through regulation of the miR-9/BRCA1/DNAJB6/KLF4/AKT1 axis in ovarian cancer. Mol Cancer 18(1):144. https://doi.org/10.1186/s12943-019-1080-5
Tang Q, Hann SS (2020) Biological roles and mechanisms of circular RNA in human cancers. Onco Targets Therap 13:2067
Ren W, Yuan Y, Peng J, Mutti L, Jiang X (2022) The function and clinical implication of circular RNAs in lung cancer. Front Oncol 12:862602. https://doi.org/10.3389/fonc.2022.862602
Dai C, Liu B, Li S, Hong Y, Si J, Xiong Y et al (2021) Construction of a circRNA-miRNA-mRNA regulated pathway involved in EGFR-TKI lung adenocarcinoma resistance. Technol Cancer Res Treat 20:15330338211056808. https://doi.org/10.1177/15330338211056809
Zepeda-Enríquez P, Silva-Cázares MB, López-Camarillo C (2023) Novel insights into circular RNAs in metastasis in breast cancer: an update. Noncoding RNA. https://doi.org/10.3390/ncrna9050055
Li Z, Chen Z, Hu G, Jiang Y (2019) Roles of circular RNA in breast cancer: present and future. Am J Transl Res 11(7):3945–3954
Gopikrishnan M, Hephzibah Cathryn R, Gnanasambandan R, Ashour HM, Pintus G, Hammad M et al (2023) Therapeutic and diagnostic applications of exosomal circRNAs in breast cancer. Funct Integr Genom 23(2):184. https://doi.org/10.1007/s10142-023-01083-3
Li X, Shen M (2019) Circular RNA hsa_circ_103809 suppresses hepatocellular carcinoma proliferation and invasion by sponging miR-620. Eur Rev Med Pharmacol Sci 23(2):555–566. https://doi.org/10.26355/eurrev_201902_16868
Panda AC, De S, Grammatikakis I, Munk R, Yang X, Piao Y et al (2017) High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs. Nucleic Acids Res 45(12):e116
Zeng X, Lin W, Guo M, Zou Q (2017) A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput Biol 13(6):e1005420
Jakobi T, Dieterich C (2019) Computational approaches for circular RNA analysis. Wiley Interdiscip Rev 10(3):e1528
Hansen TB (2018) Improved circRNA identification by combining prediction algorithms. Front Cell Dev Biol 6:20
Chen I, Chen CY, Chuang TJ (2015) Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev 6(5):563–579
Meng X, Li X, Zhang P, Wang J, Zhou Y, Chen M (2017) Circular RNA: an emerging key player in RNA world. Brief Bioinfo 18(4):547–557
Jeck WR, Sharpless NE (2014) Detecting and characterizing circular RNAs. Nat Biotechnol 32(453):461
Li HJ (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Genomics. https://doi.org/10.48550/arXiv.1303.3997
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S et al (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1):15–21
Kim D, Salzberg S (2011) TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol 12:72
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25(9):1105–1111
Cheng J, Metge F, Dieterich C (2016) Specific identification and quantification of circular RNAs from sequencing data. Bioinformatics 32(7):1094–1096
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9(4):357–359
Song X, Zhang N, Han P, Moon BS, Lai RK, Wang K et al (2016) Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res 44(9):e87
Izuogu OG, Alhasan AA, Alafghani HM, Santibanez-Koref M, Elliott DJ, Jackson MS (2016) PTESFinder: a computational method to identify post-transcriptional exon shuffling (PTES) events. Nucleic Acids Res 17(1):1–11
Szabo L, Morey R, Palpant N, Wang P, Afari N, Jiang C, Parast MM, Murry CE, Laurent LC, Salzman J (2015) Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol 16:126
Gao Y, Wang J, Zhao F (2015) CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol 16(1):1–16
Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL et al (2010) MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res 38(18):e178
Hoffmann S, Otto C, Doose G, Tanzer A, Langenberger D, Christ S et al (2014) A multi-split mapping algorithm for circular RNA, splicing, trans-splicing and fusion detection. Genome Biol 15(2):1–11
Chuang TJ, Wu CS, Chen CY, Hung LY, Chiang TW, Yang MY (2016) NCLscan: accurate identification of non-co-linear transcripts (fusion, trans-splicing and circular RNA) with a good balance between sensitivity and precision. Nucleic Acids Res 44(3):e29
Chen B, Wei W, Huang X, Xie X, Kong Y, Dai D et al (2018) circEPSTI1 as a prognostic marker and mediator of triple-negative breast cancer progression. Theranostics 8(14):4003
Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S et al (2017) Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol 14(3):361–369
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B et al (2016) Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 7:11215
Dudekula DB, Panda AC, Grammatikakis I, Supriya D, Abdelmohsen K, Gorospe M (2016) CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol 13(1):34–42
Panda AC, Abdelmohsen K, Gorospe M (2017) RT-qPCR detection of senescence-associated circular RNAs. In: Nikiforov MA (ed) Oncogene-induced senescence. Springer, New York, pp 79–87
Quan PL, Sauzade M, Brouzes EJS (2018) dPCR: a technology review. Sensors 18(4):1271
Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ et al (2011) High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 83(22):8604–8610
Zhang Y, Wang Y, Su X, Wang P, Lin W (2021) The value of circulating circular RNA in cancer diagnosis, monitoring, prognosis, and guiding treatment. Front Oncol 11:736546. https://doi.org/10.3389/fonc.2021.736546
Wang S, Zhang K, Tan S, Xin J, Yuan Q, Xu H et al (2021) Circular RNAs in body fluids as cancer biomarkers: the new frontier of liquid biopsies. Mol Cancer 20(1):13. https://doi.org/10.1186/s12943-020-01298-z
Xu CY, Zeng XX, Xu LF, Liu M, Zhang F (2022) Circular RNAs as diagnostic biomarkers for gastric cancer: a comprehensive update from emerging functions to clinical significances. Front Genet. https://doi.org/10.3389/fgene.2022.1037120
Artemaki PI, Scorilas A, Kontos CK (2020) Circular RNAs: a new piece in the colorectal cancer puzzle. Cancers (Basel). https://doi.org/10.3390/cancers12092464
De Palma FDE, Salvatore F, Pol JG, Kroemer G, Maiuri MC (2022) Circular RNAs as potential biomarkers in breast cancer. Biomedicines. https://doi.org/10.3390/biomedicines10030725
Santini D, Botticelli A, Galvano A, Iuliani M, Incorvaia L, Gristina V et al (2023) Network approach in liquidomics landscape. J Exp Clin Cancer Res 42(1):193. https://doi.org/10.1186/s13046-023-02743-9
Souza VGP, Forder A, Brockley LJ, Pewarchuk ME, Telkar N, de Araújo RP et al (2023) Liquid biopsy in lung cancer: biomarkers for the management of recurrence and metastasis. Int J Mol Sci 24(10):8894
Szczepaniak A, Bronisz A, Godlewski J (2023) Circular RNAs-new kids on the block in cancer pathophysiology and management. Cells. https://doi.org/10.3390/cells12040552
Lu K, Fan Q, Zou X (2022) Antisense oligonucleotide is a promising intervention for liver diseases. Front Pharmacol 13:1061842. https://doi.org/10.3389/fphar.2022.1061842
Li F, Yang Q, He AT, Yang BB (2021) Circular RNAs in cancer: limitations in functional studies and diagnostic potential. Semin Cancer Biol 75:49–61. https://doi.org/10.1016/j.semcancer.2020.10.002
Kristensen LS, Hansen TB, Venø MT, Kjems J (2018) Circular RNAs in cancer: opportunities and challenges in the field. Oncogene 37(5):555–565. https://doi.org/10.1038/onc.2017.361
Drula R, Braicu C, Chira S, Berindan-Neagoe I (2023) Investigating circular RNAs using qRT-PCR; roundup of optimization and processing steps. Int J Mol Sci 24(6):5721
Gharib E, Nasrabadi PN, Robichaud GA (2023) Circular RNA expression signatures provide promising diagnostic and therapeutic biomarkers for chronic lymphocytic leukemia. Cancers (Basel). https://doi.org/10.3390/cancers15051554
Tang X, Ren H, Guo M, Qian J, Yang Y, Gu C (2021) Review on circular RNAs and new insights into their roles in cancer. Comput Struct Biotechnol J 19:910–928. https://doi.org/10.1016/j.csbj.2021.01.018
Vromman M, Anckaert J, Bortoluzzi S, Buratin A, Chen CY, Chu Q et al (2023) Large-scale benchmarking of circRNA detection tools reveals large differences in sensitivity but not in precision. Nat Methods 20(8):1159–1169. https://doi.org/10.1038/s41592-023-01944-6
Cochran KR, Gorospe M, De S (2022) Bioinformatic Analysis of CircRNA from RNA-seq Datasets. Methods Mol Biol 2399:9–19. https://doi.org/10.1007/978-1-0716-1831-8_2
Acknowledgements
The authors would like to express their gratitude to Dr. Irina Zamfir, MD, RCP London, Basildon University Hospital UK, for providing professional English editing of this manuscript and for editorial support.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis, and interpretation, or in all these areas that is, revising or critically reviewing the article; giving final approval of the version to be published; agreeing on the journal to which the article has been submitted; and confirming to be accountable for all aspects of the work. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors wish to confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
Ethical approval
Not applicable.
Informed consent
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Saleem, A., Khan, M.U., Zahid, T. et al. Biological role and regulation of circular RNA as an emerging biomarker and potential therapeutic target for cancer. Mol Biol Rep 51, 296 (2024). https://doi.org/10.1007/s11033-024-09211-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11033-024-09211-3