Abstract
Heavy and irresponsible use of antibiotics in the last century has put selection pressure on the microbes to evolve even faster and develop more resilient strains. In the confrontation with such sometimes called “superbugs”, the search for new sources of biochemical antibiotics seems to have reached the limit. In the last two decades, bioactive antimicrobial peptides (AMPs), which are polypeptide chains with less than 100 amino acids, have attracted the attention of many in the control of microbial pathogens, more than the other types of antibiotics. AMPs are groups of components involved in the immune response of many living organisms, and have come to light as new frontiers in fighting with microbes. AMPs are generally produced in minute amounts within organisms; therefore, to address the market, they have to be either produced on a large scale through recombinant DNA technology or to be synthesized via chemical methods. Here, heterologous expression of AMPs within bacterial, fungal, yeast, plants, and insect cells, and points that need to be considered towards their industrialization will be reviewed.
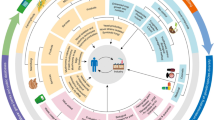
Graphical Abstract
Sources of peptide production and their applications. Some AMPs directly extracted from natural sources, some of them are chemically synthesized either using liquid or solid phase peptides synthesis, and for large scale production, recombinant expression using heterologous expression systems have been used.




Similar content being viewed by others
References
Farrokhi N, Whitelegge JP, Brusslan JA (2008) Plant peptides and peptidomics. Plant Biotechnol J 6(2):105–134
Ezzati-Tabrizi R et al (2013) Insect inducible antimicrobial peptides and their applications. Curr Protein Pept Sci 14(8):698–710
Mora C et al (2022) Over half of known human pathogenic diseases can be aggravated by climate change. Nat Clim Chang 12(9):869–875
Papadopoulos T, Baltas KN, Balta ME (2020) The use of digital technologies by small and medium enterprises during COVID-19: Implications for theory and practice. Int J Inf Manage 55:102192
Huang X et al (2020) Acinetobacter venetianus, a potential pathogen of red leg disease in freshwater-cultured whiteleg shrimp Penaeus vannamei. Aquac Rep 18:100543
Ali E et al (2021) Extract of neem (Azadirachta indica) leaf exhibits bactericidal effect against multidrug resistant pathogenic bacteria of poultry. Vet Med Sci 7(5):1921–1927
Fünfhaus A, Ebeling J, Genersch E (2018) Bacterial pathogens of bees. Curr Opin Insect Sci 26:89–96
Weldearegay YB et al (2019) Host-pathogen interactions of Mycoplasma mycoides in caprine and bovine precision-cut lung slices (PCLS) models. Pathogens 8(2):82
Algammal AM et al (2020) Emerging MDR-Pseudomonas aeruginosa in fish commonly harbor oprL and toxA virulence genes and blaTEM, blaCTX-M, and tetA antibiotic-resistance genes. Sci Rep 10(1):1–12
Ngalimat MS et al (2021) Plant growth-promoting bacteria as an emerging tool to manage bacterial rice pathogens. Microorganisms 9(4):682
Sundin GW, Wang N (2018) Antibiotic resistance in plant-pathogenic bacteria. Annu Rev Phytopathol 56:161–180
Ferri M et al (2017) Antimicrobial resistance: a global emerging threat to public health systems. Crit Rev Food Sci Nutr 57(13):2857–2876
Huan Y et al (2020) Antimicrobial peptides: classification, design, application and research progress in multiple fields. Front Microbiol. https://doi.org/10.3389/fmicb.2020.582779
Luong HX, Thanh TT, Tran TH (2020) Antimicrobial peptides - Advances in development of therapeutic applications. Life Sci 260:118407
Huang K-Y et al (2017) Identification of natural antimicrobial peptides from bacteria through metagenomic and metatranscriptomic analysis of high-throughput transcriptome data of Taiwanese oolong teas. BMC Syst Biol 11(7):29–44
Onime LA et al (2021) The rumen eukaryotome is a source of novel antimicrobial peptides with therapeutic potential. BMC Microbiol 21(1):1–13
Mishra S et al (2021) Exploiting endophytic microbes as micro-factories for plant secondary metabolite production. Appl Microbiol Biotechnol 105(18):6579–6596
Huang Y et al (2022) Isolation of lipopeptide antibiotics from Bacillus siamensis: a potential biocontrol agent for Fusarium graminearum. Can J Microbiol 99(999):1–9
Ben Khedher M et al (2022) Application and challenge of 3rd generation sequencing for clinical bacterial studies. Int J Mol Sci 23(3):1395
van der Does AM et al (2012) The human lactoferrin-derived peptide hLF1-11 exerts immunomodulatory effects by specific inhibition of myeloperoxidase activity. J Immunol 188(10):5012–5019
Mahlapuu M et al (2021) Evaluation of LL-37 in healing of hard-to-heal venous leg ulcers: a multicentric prospective randomized placebo-controlled clinical trial. Wound Repair Regen 29(6):938–950
Huang Y et al (2021) Recombinant expression of antimicrobial peptides in Pichia pastoris: a strategy to inhibit the Penicillium expansum in pears. Postharvest Biol Technol 171:111298
Shwaiki LN, Lynch KM, Arendt EK (2021) Future of antimicrobial peptides derived from plants in food application–A focus on synthetic peptides. Trends Food Sci Technol 112:312–324
Meneguetti BT et al (2017) Antimicrobial peptides from fruits and their potential use as biotechnological tools—a review and outlook. Front Microbiol 7:2136
Simons A, Alhanout K, Duval RE (2020) Bacteriocins, antimicrobial peptides from bacterial origin: Overview of their biology and their impact against multidrug-resistant bacteria. Microorganisms 8(5):639
Jiang H, Li P, Gu Q (2016) Heterologous expression and purification of plantaricin NC8, a two-peptide bacteriocin against Salmonella spp. from Lactobacillus plantarum ZJ316. Prot Exp Purif 127:28–34
Zhang J et al (2011) Expression of plectasin in Pichia pastoris and its characterization as a new antimicrobial peptide against staphyloccocus and streptococcus. Protein Expr Purif 78(2):189–196
Dotson BR et al (2018) The antibiotic peptaibol alamethicin from Trichoderma permeabilises Arabidopsis root apical meristem and epidermis but is antagonised by cellulase-induced resistance to alamethicin. BMC Plant Biol 18(1):1–11
Shenkarev ZO et al (2012) Recombinant expression and solution structure of antimicrobial peptide aurelin from jellyfish Aurelia aurita. Biochem Biophys Res Commun 429(1–2):63–69
Srinivasulu B et al (2008) Expression, purification and structural characterization of recombinant hepcidin, an antimicrobial peptide identified in Japanese flounder Paralichthys olivaceus. Prot Exp Purif 61(1):36–44
Li L et al (2005) High level expression, purification, and characterization of the shrimp antimicrobial peptide, Ch-penaeidin Pichia pastoris. Prot Exp Purif 39(2):144–151
Peng H et al (2010) Soluble expression and purification of a crab antimicrobial peptide scygonadin in different expression plasmids and analysis of its antimicrobial activity. Protein Expr Purif 70(1):109–115
Song D et al (2014) Heterologous expression and purification of dermaseptin S4 fusion in Escherichia coli and recovery of biological activity. Prep Biochem Biotechnol 44(6):598–607
Barksdale SM et al (2016) Peptides from American alligator plasma are antimicrobial against multi-drug resistant bacterial pathogens including Acinetobacter baumannii. BMC Microbiol 16(1):1–14
Xing LW et al (2016) Recombinant expression and biological characterization of the antimicrobial peptide fowlicidin-2 in Pichia pastoris. Exp Ther Med 12(4):2324–2330
Boonpa K et al (2018) Heterologous expression and antimicrobial activity of OsGASR3 from rice (Oryza sativa L.). J Plant Physiol 224:95–102
Wang H, Zhao X, Lu F (2007) Heterologous expression of bovine lactoferricin in Pichia methanolica. Biochem Mosc 72(6):640–643
Xu Z et al (2006) High-level expression of a soluble functional antimicrobial Peptide, human β-defensin 2 Escherichia coli. Biotechnol Prog 22(2):382–386
Nesa J et al (2020) Antimicrobial peptides from Bombyx mori: a splendid immune defense response in silkworms. RSC Adv 10(1):512–523
Wu Q, Patočka J, Kuča K (2018) Insect antimicrobial peptides, a mini review. Toxins. https://doi.org/10.3390/toxins10110461
Chung C-R et al (2020) Characterization and identification of natural antimicrobial peptides on different organisms. Int J Mol Sci 21(3):986
Kordi M et al (2022) Antimicrobial peptides with anticancer activity: today status, trends and their computational design. Arch Biochem Biophys 733:109484
Deb PK et al (2019) Protein/peptide drug delivery systems: practical considerations in pharmaceutical product development. Basic Fundamentals of Drug Delivery. Elsevier, pp 651–684
Akbarian M et al (2022) Bioactive peptides: synthesis, sources, applications, and proposed mechanisms of action. Int J Mol Sci 23(3):1445
Wang Y et al (2014) Influence of promoter and signal peptide on the expression of pullulanase in Bacillus subtilis. Biotech Lett 36(9):1783–1789
Ge R et al (2022) Machine learning for peptide structure, function, and design. Front Genet 13:1007635
Lei J et al (2019) The antimicrobial peptides and their potential clinical applications. Am J Transl Res 11(7):3919–3931
Koehbach J, Craik DJ (2019) The vast structural diversity of antimicrobial peptides. Trends Pharmacol Sci 40(7):517–528
Tao H et al (2022) Efficient 3D conformer generation of cyclic peptides formed by a disulfide bond. J Cheminf 14(1):26
Papagianni M (2003) Ribosomally synthesized peptides with antimicrobial properties: biosynthesis, structure, function, and applications. Biotechnol Adv 21(6):465–499
Finking R, Marahiel MA (2004) Biosynthesis of nonribosomal peptides. Annu Rev Microbiol 58(1):453–488
Walsh CT, O’Brien RV, Khosla C (2013) Nonproteinogenic amino acid building blocks for nonribosomal peptide and hybrid polyketide scaffolds. Angew Chem Int Ed 52(28):7098–7124
Hancock REW, Sahl H-G (2006) Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol 24(12):1551–1557
Rončević T, Puizina J, Tossi A (2019) Antimicrobial peptides as anti-infective agents in pre-post-antibiotic era? Int J Mol Sci. https://doi.org/10.3390/ijms20225713
Koo HB, Seo J (2019) Antimicrobial peptides under clinical investigation. Pept Sci 111(5):e24122
Wang G, Li X, Wang Z (2016) APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res 44(D1):D1087–D1093
Novković M et al (2012) DADP: the database of anuran defense peptides. Bioinformatics 28(10):1406–1407
Pirtskhalava M et al (2021) DBAASP v3: database of antimicrobial/cytotoxic activity and structure of peptides as a resource for development of new therapeutics. Nucleic Acids Res 49(D1):D288–D297
Kang X et al (2019) DRAMP 2.0, an updated data repository of antimicrobial peptides. Sci Data 6(1):148
Jhong J-H et al (2019) dbAMP: an integrated resource for exploring antimicrobial peptides with functional activities and physicochemical properties on transcriptome and proteome data. Nucleic Acids Res 47(D1):D285–D297
Gueguen Y et al (2006) PenBase, the shrimp antimicrobial peptide penaeidin database: sequence-based classification and recommended nomenclature. Dev Comp Immunol 30(3):283–288
Hammami R et al (2009) PhytAMP: a database dedicated to antimicrobial plant peptides. Nucleic Acids Res 37(1):D963–D968
Hammami R et al (2010) BACTIBASE second release: a database and tool platform for bacteriocin characterization. BMC Microbiol 10(1):1–5
van Heel AJ et al (2018) BAGEL4: a user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res 46(W1):W278–W281
Piotto SP et al (2012) YADAMP: yet another database of antimicrobial peptides. Int J Antimicrob Agents 39(4):346–351
Waghu FH et al (2016) CAMPR3: a database on sequences, structures and signatures of antimicrobial peptides. Nucleic Acids Res 44(D1):D1094–D1097
Gawde U et al (2023) CAMPR4: a database of natural and synthetic antimicrobial peptides. Nucleic Acids Res 51(D1):D377–D383
Gaspar D, Veiga AS, Castanho MA (2013) From antimicrobial to anticancer peptides. A Rev Front Microbiol 4:294
Lee, H.-T., et al., A large-scale structural classification of antimicrobial peptides. BioMed research international, 2015. 2015.
Zhao X et al (2013) LAMP: a database linking antimicrobial peptides. PLoS ONE 8(6):e66557
Wi C-I et al (2017) Application of a natural language processing algorithm to asthma ascertainment. An automated chart review. Am J Res Crit Care Med 196(4):430–437
Seebah S et al (2007) 2007 Defensins knowledgebase: a manually curated database and information source focused on the defensins family of antimicrobial peptides. Nucleic Acids Res 35(1):265–268
Fjell CD, Hancock RE, Cherkasov A (2007) AMPer: a database and an automated discovery tool for antimicrobial peptides. Bioinformatics 23(9):1148–1155
Pirtskhalava M et al (2016) DBAASP v. 2: an enhanced database of structure and antimicrobial/cytotoxic activity of natural and synthetic peptides. Nucleic Acids Res 44:D1104–D1112
Chu H-L et al (2015) Novel antimicrobial peptides with high anticancer activity and selectivity. PLoS ONE 10(5):e0126390
Frederix PW, Patmanidis I, Marrink SJ (2018) Molecular simulations of self-assembling bio-inspired supramolecular systems and their connection to experiments. Chem Soc Rev 47(10):3470–3489
Pant S et al (2020) Peptide-like and small-molecule inhibitors against Covid-19. J Biomol Struct Dynam 39(8):2904–2913
Palmer N et al (2021) Molecular dynamics for antimicrobial peptide discovery. Infect Immun. https://doi.org/10.1128/iai.00703-20
Phillips JC et al (2020) Scalable molecular dynamics on CPU and GPU architectures with NAMD. J Chem Phys. https://doi.org/10.1063/5.0014475
Abraham MJ et al (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1:19–25
Case DA et al (2021) Amber 2021. University of California, San Francisco
Melo MC et al (2020) Generalized correlation-based dynamical network analysis: a new high-performance approach for identifying allosteric communications in molecular dynamics trajectories. J Chem Phys. https://doi.org/10.1063/5.0018980
Das P et al (2021) Accelerated antimicrobial discovery via deep generative models and molecular dynamics simulations. Nat Biomed Eng 5(6):613–623
Ruiz Puentes P et al (2022) Rational discovery of antimicrobial peptides by means of artificial intelligence. Membranes 12(7):708
Ruiz Puentes P et al (2022) Rational discovery of antimicrobial peptides by means of artificial intelligence. Membranes (Basel) 12(7):708
Sowers A et al (2023) Advances in antimicrobial peptide discovery via machine learning and delivery via nanotechnology. Microorganisms 11(5):1129. https://doi.org/10.3390/microorganisms11051129
Lee EY et al (2017) What can machine learning do for antimicrobial peptides, and what can antimicrobial peptides do for machine learning? Interface Focus 7(6):20160153
Jhong J-H et al (2022) dbAMP 2.0: updated resource for antimicrobial peptides with an enhanced scanning method for genomic and proteomic data. Nucleic Acids Res 50:D460–D470
Lin D et al (2022) Mining amphibian and insect transcriptomes for antimicrobial peptide sequences with rAMPage. Antibiotics 11(7):952
Mirski T et al (2018) Utilisation of peptides against microbial infections–a review. Ann Agric Environ Med 25(2):205–210
Santos R et al (2016) Bacterial biofilms in diabetic foot ulcers: potential alternative therapeutics. Microbial Biofilms-Importance Appl. https://doi.org/10.5772/63085
Neshani A et al (2019) Epinecidin-1, a highly potent marine antimicrobial peptide with anticancer and immunomodulatory activities. BMC Pharmacol Toxicol 20(1):1–11
Scarsini M et al (2015) Antifungal activity of cathelicidin peptides against planktonic and biofilm cultures of Candida species isolated from vaginal infections. Peptides 71:211–221
Madanchi H et al (2020) Antimicrobial peptides of the vaginal innate immunity and their role in the fight against sexually transmitted diseases. New Microbes New Infect 34:100627
Kordi M et al (2023) Antimicrobial peptides with anticancer activity: Today status, trends and their computational design. Arch Biochem Biophys 733:109484
Chen CH, Lu TK (2020) Development and challenges of antimicrobial peptides for therapeutic applications. Antibiotics 9(1):24
Dijksteel GS et al (2021) Review: lessons learned from clinical trials using Antimicrobial Peptides (AMPs). Front Microbiol. https://doi.org/10.3389/fmicb.2021.616979
Moretta A et al (2021) Antimicrobial peptides: a new hope in biomedical and pharmaceutical fields. Front Cell Infect Microbiol 11:453
Fukuta S et al (2012) Transgenic tobacco plants expressing antimicrobial peptide bovine lactoferricin show enhanced resistance to phytopathogens. Plant Biotechnol 29(4):383–389
Khademi M et al (2020) New recombinant antimicrobial peptides confer resistance to fungal pathogens in tobacco plants. Front Plant Sci 11:1236
Chahardoli M, Fazeli A, Ghabooli M (2018) Recombinant production of bovine Lactoferrin-derived antimicrobial peptide in tobacco hairy roots expression system. Plant Physiol Biochem 123:414–421
Rai M et al (2016) Antimicrobial peptides as natural bio-preservative to enhance the shelf-life of food. J Food Sci Technol 53(9):3381–3394
Tkaczewska J (2020) Peptides and protein hydrolysates as food preservatives and bioactive components of edible films and coatings-A review. Trends Food Sci Technol 106:298–311
Jia L et al (2021) Bioactive peptides from foods: production, function, and application. Food Funct 12(16):7108–7125
Xu Y (2021) Phage and phage lysins: new era of bio-preservatives and food safety agents. J Food Sci 86(8):3349–3373
Querido MM et al (2019) Self-disinfecting surfaces and infection control. Coll Surf, B 178:8–21
Li FF, Brimble MA (2019) Using chemical synthesis to optimise antimicrobial peptides in the fight against antimicrobial resistance. Pure Appl Chem 91(2):181–198
Beutler JA (2009) Natural products as a foundation for drug discovery. Curr Protoc Pharmacol. https://doi.org/10.1002/0471141755.ph0911s46
Kim DS et al (2019) A new prokaryotic expression vector for the expression of antimicrobial peptide abaecin using SUMO fusion tag. BMC Biotechnol 19(1):1–12
Seyedjavadi SS et al (2021) Design, dimerization, and recombinant production of MCh-AMP1–derived peptide in Escherichia coli and evaluation of its antifungal activity and cytotoxicity. Front Fungal Biol 2:638595
Sinha R, Shukla P (2019) Antimicrobial peptides: recent insights on biotechnological interventions and future perspectives. Protein Pept Lett 26(2):79–87
Luiz DP et al (2017) Heterologous expression of abaecin peptide from Apis mellifera in Pichia pastoris. Microb Cell Fact 16(1):1–7
Liu Y et al (2016) Topical delivery of low-cost protein drug candidates made in chloroplasts for biofilm disruption and uptake by oral epithelial cells. Biomaterials 105:156–166
Li Z et al (2011) Expression of a radish defensin in transgenic wheat confers increased resistance to Fusarium graminearum and Rhizoctonia cerealis. Funct Integr Genom 11(1):63–70
Silva-Carvalho AÉ et al (2021) Dissecting the relationship between antimicrobial peptides and mesenchymal stem cells. Pharmacol Ther 233:108021
DMM, Jaradat (2018) Thirteen decades of peptide synthesis: key developments in solid phase peptide synthesis and amide bond formation utilized in peptide ligation. Amino Acids 50(1):39–68
Hou W, Zhang X, Liu C-F (2017) Progress in chemical synthesis of peptides and proteins. Trans Tianjin Univ 23(5):401–419
Behrendt R, White P, Offer J (2016) Advances in Fmoc solid-phase peptide synthesis. J Pept Sci 22(1):4–27
Petrou C, Sarigiannis Y (2018) Peptide synthesis: methods, trends, and challenges. Peptide Appl Biomed Biotechnol Bioeng. https://doi.org/10.1016/B978-0-08-100736-5.00001-6
Burcu, U., et al., Synthesis and Applications of Synthetic Peptides, in Peptide Synthesis, T.V. Jaya, Editor. 2019, IntechOpen: Rijeka
Fischer PM, Zheleva DI (2002) Liquid-phase peptide synthesis on polyethylene glycol (PEG) supports using strategies based on the 9-fluorenylmethoxycarbonyl amino protecting group: application of PEGylated peptides in biochemical assays. J Peptide Sci: Off Publ Eur Peptide Soc 8(9):529–542
Martin V et al (2020) Greening the synthesis of peptide therapeutics: an industrial perspective. RSC Adv 10(69):42457–42492
Yeo JIN (2021) Liquid phase peptide synthesis via nanostar sieving. Angew Chem 133(14):7865–7874
Chan W, White P (1999) Fmoc Solid Phase Peptide Synthesis: A Practical Approach. Oxford University Press, Oxford
Isidro-Llobet A et al (2019) Sustainability challenges in peptide synthesis and purification: from R&D to production. J Org Chem 84(8):4615–4628
Mäde V, Els-Heindl S, Beck-Sickinger AG (2014) Automated solid-phase peptide synthesis to obtain therapeutic peptides. Beilstein J Org Chem 10(1):1197–1212
Li W et al (2021) Chemically modified and conjugated antimicrobial peptides against superbugs. Chem Soc Rev 50(8):4932–4973
Guo L et al (2023) Lipidated variants of the antimicrobial peptide nisin produced via incorporation of methionine analogs for click chemistry show improved bioactivity. J Biol Chem 299(7):104845
Roscetto E et al (2021) Antimicrobial activity of a lipidated Temporin L analogue against carbapenemase-producing klebsiella pneumoniae clinical isolates. Antibiotics 10(11):1312
Grimsey E et al (2020) 2020 The effect of lipidation and glycosylation on short cationic antimicrobial peptides. Biomembranes 8:183195
Deo S et al (2022) Strategies for improving antimicrobial peptide production. Biotechnol Adv 59:107968
Wang L et al (2022) Therapeutic peptides: current applications and future directions. Signal Transduct Target Ther 7(1):1–27
Kordi, M., et al., Peptide production by molecular farming with antiviral effects. Agricultural Bioeconomy : Innovation and Foresight in the Post-COVID Era, 2022.
Lou Y-C et al (2006) Roles of N-terminal pyroglutamate in maintaining structural integrity and pKa values of catalytic histidine residues in bullfrog ribonuclease 3. J Mol Biol 355(3):409–421
Moerman P (2014) Novel methods for C-terminal sequence analysis in the proteome era. Ghent University, Ghent
Prokai-Tatrai K (2012) Modifying peptide properties by prodrug design for. Peptide Transport Deliv Central Nerv Syst 61:155
Kuzmin D et al (2017) Effect of N-and C-terminal modifications on cytotoxic properties of antimicrobial peptide tachyplesin I. Bull Exp Biol Med 162(6):754–757
Romo TD et al (1808) 2011 Membrane binding of an acyl-lactoferricin B antimicrobial peptide from solid-state NMR experiments and molecular dynamics simulations. Biochimica et Biophysica Acta (BBA) - Biomembr 8:2019–2030
Uzzell T et al (2003) Hagfish intestinal antimicrobial peptides are ancient cathelicidins. Peptides 24(11):1655–1667
Tasiemski A et al (2007) Hedistin: a novel antimicrobial peptide containing bromotryptophan constitutively expressed in the NK cells-like of the marine annelid. Nereis Diversicolor Develop Comp Immunol 31(8):749–762
Castiglione F et al (2008) Determining the structure and mode of action of microbisporicin, a potent lantibiotic active against multiresistant pathogens. Chem Biol 15(1):22–31
Wu M-H et al (2020) Effects of glycosylation and D-amino acid substitution on the antitumor and antibacterial activities of bee venom peptide HYL. Bioconjug Chem 31(10):2293–2302
Lohans CT, Vederas JC (2014) Structural characterization of thioether-bridged bacteriocins. J Antibiot 67(1):23–30
Vernen F et al (2019) Characterization of Tachyplesin peptides and their cyclized analogues to improve antimicrobial and anticancer properties. Int J Mol Sci. https://doi.org/10.3390/ijms20174184
Wang G (2012) Post-translational modifications of natural antimicrobial peptides and strategies for peptide engineering. Curr Biotechnol 1(1):72–79
Li D et al (2021) N-terminal acetylation of antimicrobial peptide L163 improves its stability against protease degradation. J Pept Sci 27(9):e3337
Xu J et al (2012) Green factory: plants as bioproduction platforms for recombinant proteins. Biotechnol Adv 30(5):1171–1184
Cohen P (2001) The role of protein phosphorylation in human health and disease. Eur J Biochem 268(19):5001–5010
Tan CSH et al (2009) Comparative analysis reveals conserved protein phosphorylation networks implicated in multiple diseases. Sci Signal 2(81):ra39–ra39
Meinnel T, Giglione C (2008) Protein lipidation meets proteomics. Front Biosci-Landmark 13(16):6326–6340
Glenz K et al (2006) Production of a recombinant bacterial lipoprotein in higher plant chloroplasts. Nat Biotechnol 24(1):76–77
Chen P et al (2023) Embracing the era of antimicrobial peptides with marine organisms. Nat Product Rep. https://doi.org/10.1039/D3NP00031A
Hou C et al (2014) Production of antibacterial peptide from bee venom via a new strategy for heterologous expression. Mol Biol Rep 41(12):8081–8091
Thomas X et al (2004) Siderophore peptide, a new type of post-translationally modified antibacterial peptide with potent activity. J Biol Chem 279(27):28233–28242
Balducci E et al (2011) Structural and functional consequences induced by post-translational modifications in <i>α</i>-Defensins. Int J Peptides 2011:594723
Zheng Q, Fang H, Liu W (2017) Post-translational modifications involved in the biosynthesis of thiopeptide antibiotics. Org Biomol Chem 15(16):3376–3390
Ingham AB, Moore RJ (2007) Recombinant production of antimicrobial peptides in heterologous microbial systems. Biotechnol Appl Biochem 47(1):1–9
Ko H et al (2021) A novel protein fusion partner, carbohydrate-binding module family 66, to enhance heterologous protein expression in Escherichia coli. Microb Cell Fact 20:1–12
Lamer T et al (2022) SPI “Sandwich”: combined SUMO-peptide-Intein expression system and isolation procedure for improved stability and yield of peptides. Protein Sci 31(5):e4316
Pane K et al (2016) Rational design of a carrier protein for the production of recombinant toxic peptides in Escherichia coli. PLoS ONE 11(1):e0146552
Goel A et al (2000) Relative position of the hexahistidine tag effects binding properties of a tumor-associated single-chain Fv construct. Gen Subj 1523(1):13–20
Cantrell MS et al (2022) Expression and purification of a cleavable recombinant fortilin from Escherichia coli for structure activity studies. Protein Expr Purif 189:105989
Cipakova I, Hostinová E (2005) Production of the human-beta-defensin using Saccharomyces cerevisiae as a host. Protein Pept Lett 12(6):551–554
Ishizaki K et al (2015) Development of gateway binary vector series with four different selection markers for the liverwort Marchantia polymorpha. PLoS ONE 10(9):e0138876
Komori T, Ueki J, Komari T (2011) Transformation vectors and expression of foreign genes in higher plants. Hist Technol Develop 1:55
Nakagawa T et al (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104(1):34–41
Chen P-Y et al (2003) Complete sequence of the binary vector pBI121 and its application in cloning T-DNA insertion from transgenic plants. Mol Breeding 11:287–293
Jia B, Jeon CO (2016) High-throughput recombinant protein expression in Escherichia coli: current status and future perspectives. Open Biol 6(8):160196
Hozjan V et al (2008) Ligand supplementation as a method to increase soluble heterologous protein production. Expert Rev Proteomics 5(1):137–143
Hong I-P et al (2007) Recombinant expression of human cathelicidin (hCAP18/LL-37) in Pichia pastoris. Biotech Lett 29(1):73–78
Quezada-Rivera J et al (2019) Heterologous expression of bacteriocin E-760 in Chlamydomonas reinhardtii and functional analysis. Phyton 88(1):25
Ishvaanjil B et al (2014) Heterologous expression of antimicrobial peptide LL-37 in Chinese cabbage with enhanced resistance to pathogens. Mong J Agric Sci 13(2):124–130
Eustáquio AS et al (2005) Heterologous expression of novobiocin and clorobiocin biosynthetic gene clusters. Appl Environ Microbiol 71(5):2452–2459
Zhang L et al (2020) Heterologous expression of the novel α-helical hybrid peptide PR-FO in Bacillus subtilis. Bioprocess Biosyst Eng 43:1619–1627
Zhang M et al (2018) Expression of a recombinant hybrid antimicrobial peptide magainin II-cecropin B in the mycelium of the medicinal fungus Cordyceps militaris and its validation in mice. Microb Cell Fact 17(1):18
Parachin NS et al (2012) Expression systems for heterologous production of antimicrobial peptides. Peptides 38(2):446–456
Shanmugaraj B et al (2021) Biotechnological insights on the expression and production of antimicrobial peptides in plants. Molecules 26(13):4032
Rosano GL, Ceccarelli EA (2014) Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol 5:172
Eichmann J et al (2019) Selection of high producers from combinatorial libraries for the production of recombinant proteins in Escherichia coli and Vibrio natriegens. Front Bioeng Biotechnol 7:254
Deng T et al (2017) The heterologous expression strategies of antimicrobial peptides in microbial systems. Protein Expr Purif 140:52–59
Hammers D, Carothers K, Lee S (2022) The role of bacterial proteases in microbe and host-microbe interactions. Curr Drug Targets 23(3):222–239
Chen CH, Lu TK (2020) Development and challenges of antimicrobial peptides for therapeutic applications. Antibiotics (Basel) 9(1):24
Li Y (2011) Recombinant production of antimicrobial peptides in Escherichia coli: a review. Protein Expr Purif 80(2):260–267
Utkina L et al (2010) Heterologous expression of a synthetic gene encoding a novel hevein-type antimicrobial peptide of Leymus arenarius in Escherichia coli cells. Russ J Genet 46:1449–1454
Chu X et al (2023) Heterologous expression and bioactivity determination of monochamus alternatus antibacterial peptide gene in Komagataella phaffii (Pichia pastoris). Int J Mol Sci 24(6):5421
Elhag O et al (2017) 2017 Screening, expression, purification and functional characterization of novel antimicrobial peptide genes from Hermetia illucens (L.). PLoS ONE 12(1):e0169582
Kawai Y et al (2003) Heterologous expression of gassericin A, a bacteriocin produced by Lactobacillus gasseri LA39. Anim Sci J 74(1):45–51
ROSLI, N., et al (2019) Heterologous expression of recombinant scygonadin antimicrobial peptide from mud crab scylla serrata. Malays Appl Biol 48(1):95–100
Maarof E et al (2011) Cloning and heterologous expression of’CDef1’, a ripening-induced defensin from’capsicum annuum’. Aust J Crop Sci 5(3):271–276
Nigutová K et al (2008) Heterologous expression of functionally active enterolysin A, class III bacteriocin from Enterococcus faecalis Escherichia coli. Prot Exp Purif 60(1):20–24
Ram KS et al (2014) Cost effective purification of intein based syntetic cationic antimicrobial peptide expressed in cold shock expression system using salt inducible E coli GJ1158. J Microbiol Infect Dis 4(01):13–19
Meiyalaghan S et al (2014) Expression and purification of the antimicrobial peptide GSL1 in bacteria for raising antibodies. BMC Res Notes 7(1):1–7
Brede DA et al (2005) Heterologous production of antimicrobial peptides in Propionibacterium freudenreichii. Appl Environ Microbiol 71(12):8077–8084
Song D, Li P, Gu Q (2018) Cloning and heterologous expression of plantaricin ZJ5, a novel bacteriocin from Lactobacillus plantarum ZJ5, in Escherichia coli. Nat Prod Commun. https://doi.org/10.1177/1934578X1801301231
Roldan-Tapia M et al (2017) Streptomyces as overexpression system for heterologous production of an antimicrobial peptide. Protein Pept Lett 24(6):483–488
Martín M et al (2007) Cloning, production and expression of the bacteriocin enterocin A produced by Enterococcus faecium PLBC21 in Lactococcus lactis. Appl Microbiol Biotechnol 76(3):667–675
Fernández M et al (2007) Heterologous expression of enterocin AS-48 in several strains of lactic acid bacteria. J Appl Microbiol 102(5):1350–1361
Herbel V, Schäfer H, Wink M (2015) Recombinant production of snakin-2 (an antimicrobial peptide from tomato) in E. coli and analysis of its bioactivity. Molecules 20(8):14889–14901
Beaulieu L et al (2007) Production of active pediocin PA-1 in Escherichia coli using a thioredoxin gene fusion expression approach: cloning, expression, purification, and characterization. Can J Microbiol 53(11):1246–1258
Tavares LS et al (2012) Antimicrobial activity of recombinant Pg-AMP1, a glycine-rich peptide from guava seeds. Peptides 37(2):294–300
Klocke M et al (2005) Heterologous expression of enterocin A, a bacteriocin from Enterococcus faecium, fused to a cellulose-binding domain in Escherichia coli results in a functional protein with inhibitory activity against Listeria. Appl Microbiol Biotechnol 67(4):532–538
Ma D-Y et al (2008) Expression and characterization of recombinant gallinacin-9 and gallinacin-8 in Escherichia coli. Protein Expr Purif 58(2):284–291
Zhou Q-F et al (2007) High-level production of a novel antimicrobial peptide perinerin in Escherichia coli by fusion expression. Curr Microbiol 54(5):366–370
Choi HJ et al (2005) Heterologous expression of human $\beta $-Defensin-1 in bacteriocin-producing laetoeoeeus lactis. J Microbiol Biotechnol 15(2):330–336
Odintsova TI et al (2009) A novel antifungal hevein-type peptide from Triticum kiharae seeds with a unique 10-cysteine motif. FEBS J 276(15):4266–4275
Ryazantsev DY et al (2014) A novel hairpin-like antimicrobial peptide from barnyard grass (Echinochloa crusgalli L.) seeds: structure–functional and molecular-genetics characterization. Biochimie 99:63–70
Gizatullina AK et al (2013) Recombinant production and solution structure of lipid transfer protein from lentil Lens culinaris. Biochem Biophys Res Commun 439(4):427–432
Bogdanov IV et al (2016) A novel lipid transfer protein from the pea Pisum sativum: isolation, recombinant expression, solution structure, antifungal activity, lipid binding, and allergenic properties. BMC Plant Biol 16:1–17
Slavokhotova AA et al (2014) Novel antifungal α-hairpinin peptide from Stellaria media seeds: structure, biosynthesis, gene structure and evolution. Plant Mol Biol 84:189–202
Xu J et al (2020) Transgenic expression of antimicrobial peptides from black soldier fly enhance resistance against entomopathogenic bacteria in the silkworm, Bombyx mori. Insect Biochem Mol Biol 127:103487
Vandermies M, Fickers P (2019) Bioreactor-scale strategies for the production of recombinant protein in the yeast yarrowia lipolytica. Microorganisms 7(2):40
Madhavan A et al (2021) Customized yeast cell factories for biopharmaceuticals: from cell engineering to process scale up. Microb Cell Fact 20(1):124
Huang M et al (2017) Efficient protein production by yeast requires global tuning of metabolism. Nat Commun 8(1):1–12
Lestari C, Novientri G (2021) Advantages of yeast-based recombinant protein technology as vaccine products against infectious diseases. IOP Conf Series: Earth Environ Sci 913(1):012099
Mohammadzadeh R et al (2021) Practical methods for expression of recombinant protein in the pichia pastoris system. Curr Protoc 1(6):e155
Kant P, Liu W-Z, Pauls KP (2009) PDC1, a corn defensin peptide expressed in Escherichia coli and Pichia pastoris inhibits growth of Fusarium graminearum. Peptides 30(9):1593–1599
Ma Y, Lee C-J, Park J-S (2020) Strategies for optimizing the production of proteins and peptides with multiple disulfide bonds. Antibiotics 9(9):541
Vieira Gomes AM et al (2018) Comparison of yeasts as hosts for recombinant protein production. Microorganisms 6(2):38
Liu Z et al (2012) Different expression systems for production of recombinant proteins in Saccharomyces cerevisiae. Biotechnol Bioeng 109(5):1259–1268
Zhao H et al (2015) Characterization of bioactive recombinant antimicrobial peptide parasin I fused with human lysozyme expressed in the yeast Pichia pastoris system. Enzyme Microb Technol 77:61–67
Meng D-M et al (2016) Expression, purification and initial characterization of a novel recombinant antimicrobial peptide Mytichitin-A in Pichia pastoris. Protein Expr Purif 127:35–43
Chen Z et al (2011) Recombinant antimicrobial peptide hPAB-β expressed in Pichia pastoris, a potential agent active against methicillin-resistant Staphylococcus aureus. Appl Microbiol Biotechnol 89(2):281–291
Jin F-L et al (2009) Expression and characterization of antimicrobial peptide CecropinAD in the methylotrophic yeast Pichia pastoris. Process Biochem 44(1):11–16
Kuddus MR et al (2016) Expression, purification and characterization of the recombinant cysteine-rich antimicrobial peptide snakin-1 in Pichia pastoris. Protein Expr Purif 122:15–22
Chen X et al (2017) High-level heterologous production and Functional Secretion by recombinant Pichia pastoris of the shortest proline-rich antibacterial honeybee peptide Apidaecin. Sci Rep 7(1):1–9
Van Reenen C et al (2003) Characterization and heterologous expression of a class IIa bacteriocin, plantaricin 423 from Lactobacillus plantarum 423, in Saccharomyces cerevisiae. Int J Food Microbiol 81(1):29–40
Song X et al (2005) cDNA cloning, functional expression and antifungal activities of a dimeric plant defensin SPE10 from Pachyrrhizus erosus seeds. Plant Mol Biol 57(1):13–20
Vasil IK (2008) A history of plant biotechnology: from the cell theory of Schleiden and Schwann to biotech crops. Plant Cell Rep 27(9):1423–1440
Abdulhafiz F (2022) Plant cell culture technologies: a promising alternatives to produce high-value secondary metabolites. Arab J Chem 15(11):104161
Ghidey M et al (2020) Making plants into cost-effective bioreactors for highly active antimicrobial peptides. New Biotechnol 56:63–70
Bakare OO et al (2022) Plant antimicrobial peptides (PAMPs): features, applications, production, expression, and challenges. Molecules 27(12):3703
Kordi M et al (2022) Peptide production by molecular farming with antiviral effects. Agricultural Bioeconomy. Elsevier
Goyal RK et al (2013) Expression of an engineered heterologous antimicrobial peptide in potato alters plant development and mitigates normal abiotic and biotic responses. PLoS ONE 8(10):e77505
Lacerda AF et al (2014) Antifungal defensins and their role in plant defense. Front Microbiol 5:116
Kumar M, Kumar V, Prasad R (2020) Phyto-microbiome in stress regulation. Springer, Singapore
Burnett MJ, Burnett AC (2020) Therapeutic recombinant protein production in plants: Challenges and opportunities. Plants, People, Planet 2(2):121–132
Gleba Y, Klimyuk V, Marillonnet S (2005) Magnifection—a new platform for expressing recombinant vaccines in plants. Vaccine 23(17–18):2042–2048
Patiño-Rodríguez O et al (2013) Transient expression and characterization of the antimicrobial peptide protegrin-1 in Nicotiana tabacum for control of bacterial and fungal mammalian pathogens. Plant Cell. Tissue Organ Cult (PCTOC) 115:99–106
Chaudhary S et al (2023) Efficient in planta production of amidated antimicrobial peptides that are active against drug-resistant ESKAPE pathogens. Nat Commun 14(1):1464
Campos-Quevedo N et al (2013) Production of milk-derived bioactive peptides as precursor chimeric proteins in chloroplasts of Chlamydomonas reinhardtii. Plant Cell. Tissue and Organ Cult (PCTOC) 113(2):217–225
Koo J, Park D, Kim H (2013) Expression of bovine lactoferrin N-lobe by the green alga. Chlorella vulgaris Algae 28(4):379–387
Wang K et al (2020) Chloroplast genetic engineering of a unicellular green alga Haematococcus pluvialis with expression of an antimicrobial peptide. Mar Biotechnol 22(4):572–580
Rosales-Mendoza S, Paz-Maldonado LMT, Soria-Guerra RE (2012) Chlamydomonas reinhardtii as a viable platform for the production of recombinant proteins: current status and perspectives. Plant Cell Rep 31(3):479–494
Barbosa Viana AA, Pelegrini PB, Grossi-de-Sá MF (2012) Plant biofarming: novel insights for peptide expression in heterologous systems. Pept Sci 98(4):416–427
Mirzaee M et al (2021) Long-lasting stable expression of human LL-37 Antimicrobial peptide in transgenic barley plants. Antibiotics 10(8):898
Decker EL, Reski R (2004) The moss bioreactor. Curr Opin Plant Biol 7(2):166–170
Terrier B et al (2007) Two new disposable bioreactors for plant cell culture: the wave and undertow bioreactor and the slug bubble bioreactor. Biotechnol Bioeng 96(5):914–923
Decker EL, Reski R (2008) Current achievements in the production of complex biopharmaceuticals with moss bioreactors. Bioprocess Biosyst Eng 31(1):3–9
Ducos J-P, Terrier B, Courtois D (2009) Disposable bioreactors for plant micropropagation and mass plant cell culture. In: Eibl R, Eibl D (eds) Disposable bioreactors. Springer, Berlin
Verma D, Daniell H (2007) Chloroplast vector systems for biotechnology applications. Plant Physiol 145(4):1129–1143
Han S et al (2022) Two foreign antimicrobial peptides expressed in the chloroplast of Porphyridium purpureum possessed antibacterial properties. Mar Drugs 20(8):484
Islam S, Bezbaruah S, Kalita J (2016) A review on antimicrobial peptides from Bombyxmori L and their application in plant and animal disease control. J Adv Bio Biotechnol 9(3):2394–1081
Rahnamaeian M et al (2009) Insect peptide metchnikowin confers on barley a selective capacity for resistance to fungal ascomycetes pathogens. J Exp Bot 60(14):4105–4114
Almasia NI et al (2008) Overexpression of snakin-1 gene enhances resistance to Rhizoctonia solani and Erwinia carotovora in transgenic potato plants. Mol Plant Pathol 9(3):329–338
Hammond J et al (2006) Transgenic approaches to disease resistance in ornamental crops. J Crop Improv 17(1–2):155–210
Jung Y-J (2013) Enhanced resistance to bacterial pathogen in transgenic tomato plants expressing cathelicidin antimicrobial peptide. Biotechnol Bioprocess Eng 18(3):615–624
Jan P-S, Huang H-Y, Chen H-M (2010) Expression of a synthesized gene encoding cationic peptide cecropin B in transgenic tomato plants protects against bacterial diseases. Appl Environ Microbiol 76(3):769–775
Shi X et al (2019) Efficient production of antifungal proteins in plants using a new transient expression vector derived from tobacco mosaic virus. Plant Biotechnol J 17(6):1069–1080
DeGray G et al (2001) Expression of an antimicrobial peptide via the chloroplast genome to control phytopathogenic bacteria and fungi. Plant Physiol 127(3):852–862
Jaber E et al (2017) A gene encoding scots pine antimicrobial protein Sp-AMP2 (PR-19) confers increased tolerance against Botrytis cinerea in transgenic tobacco. Forests 9(1):10
Aerts AM et al (2007) Arabidopsis thaliana plants expressing human beta-defensin-2 are more resistant to fungal attack: functional homology between plant and human defensins. Plant Cell Rep 26(8):1391–1398
Khan RS et al (2006) Transgenic potatoes expressing wasabi defensin peptide confer partial resistance to gray mold (Botrytis cinerea). Plant biotechnology 23(2):179–183
Osusky M et al (2004) Transgenic potatoes expressing a novel cationic peptide are resistant to late blight and pink rot. Transgenic Res 13(2):181–190
Wu T et al (2013) Expression of antimicrobial peptides thanatin (S) in transgenic Arabidopsis enhanced resistance to phytopathogenic fungi and bacteria. Gene 527(1):235–242
Rivero M et al (2012) Stacking of antimicrobial genes in potato transgenic plants confers increased resistance to bacterial and fungal pathogens. J Biotechnol 157(2):334–343
Rahnamaeian M, Vilcinskas A (2012) Defense gene expression is potentiated in transgenic barley expressing antifungal peptide metchnikowin throughout powdery mildew challenge. J Plant Res 125(1):115–124
Muramoto N et al (2012) Transgenic sweet potato expressing thionin from barley gives resistance to black rot disease caused by Ceratocystis fimbriata in leaves and storage roots. Plant Cell Rep 31(6):987–997
Zhou M et al (2011) Expression of a novel antimicrobial peptide Penaeidin4–1 in creeping bentgrass (Agrostis stolonifera L.) enhances plant fungal disease resistance. PLoS ONE 6(9):e24677
Krens FA et al (2011) Performance and long-term stability of the barley hordothionin gene in multiple transgenic apple lines. Transgenic Res 20(5):1113–1123
Ntui VO et al (2010) Stable integration and expression of wasabi defensin gene in “Egusi” melon (Colocynthis citrullus L.) confers resistance to Fusarium wilt and Alternaria leaf spot. Plant Cell Rep 29(9):943–954
Choi M-S et al (2009) Expression of Br D1, a plant defensin from Brassica rapa, confers resistance against brown planthopper (Nilaparvata lugens) in transgenic rices. Mol Cells 28(2):131–137
Li S et al (2008) Transient expression of chicken alpha interferon gene in lettuce. J Zhejiang Univ Sci B 9(5):351–355
Yevtushenko DP, Misra S (2007) Comparison of pathogen-induced expression and efficacy of two amphibian antimicrobial peptides, MsrA2 and temporin A, for engineering wide-spectrum disease resistance in tobacco. Plant Biotechnol J 5(6):720–734
Weber W, Fussenegger M (2009) Insect cell-based recombinant protein production. Cell and tissue reaction engineering. Springer, pp 263–277
Vilcinskas A (2013) Yellow Biotechnology IIInsect Biotechnology in Plant Protectionand Industry. Springer, Berlin
Vilcinskas A (2011) Anti-infective therapeutics from the Lepidopteran model host Galleria mellonella. Curr Pharm Des 17(13):1240–1245
Käßer L et al (2022) The effect of different insect cell culture media on the efficiency of protein production by Spodoptera frugiperda cells. Electron J Biotechnol 56:54–64
Zitzmann J et al (2017) Process optimization for recombinant protein expression in insect cells. In: Gowder SJT (ed) New insights into cell culture technology. InTech, London, pp 43–97
Shu B et al (2017) Transcriptome analysis of Spodoptera frugiperda Sf9 cells reveals putative apoptosis-related genes and a preliminary apoptosis mechanism induced by azadirachtin. Sci Rep 7(1):1–13
Lemaitre RP et al (2019) FlexiBAC: a versatile, open-source baculovirus vector system for protein expression, secretion, and proteolytic processing. BMC Biotechnol 19(1):1–11
Harrison RL, Jarvis DL (2007) Transforming lepidopteran insect cells for continuous recombinant protein expression. Baculovirus and Insect Cell Expression Protocols. Springer, Berlin, pp 299–315
Valore EV et al (1998) Human beta-defensin-1: an antimicrobial peptide of urogenital tissues. J Clin Investig 101(8):1633–1642
Nakamura I et al (2001) Production of Recombinant Bovine Lactoferrin N-lobe in insect cells and its antimicrobial activity. Protein Expr Purif 21(3):424–431
Almasia NI et al (2017) Successful production of the potato antimicrobial peptide Snakin-1 in baculovirus-infected insect cells and development of specific antibodies. BMC Biotechnol 17(1):75
Käßer L et al (2022) Process intensification for the continuous production of an antimicrobial peptide in stably-transformed Sf-9 insect cells. Sci Rep 12(1):1086
Müller H, Salzig D, Czermak P (2015) Considerations for the process development of insect-derived antimicrobial peptide production. Biotechnol Prog 31(1):1–11
Panteleev PV, Ovchinnikova TV (2017) Improved strategy for recombinant production and purification of antimicrobial peptide tachyplesin I and its analogs with high cell selectivity. Biotechnol Appl Biochem 64(1):35–42
Mao R et al (2015) Optimization of expression conditions for a novel NZ2114-derived antimicrobial peptide-MP1102 under the control of the GAP promoter in Pichia pastoris X-33. BMC Microbiol 15(1):57
Wei Q et al (2005) Facilitation of expression and purification of an antimicrobial peptide by fusion with baculoviral polyhedrin in Escherichia coli. Appl Environ Microbiol 71(9):5038–5043
Beckert A et al (2015) Two c-type lysozymes boost the innate immune system of the invasive ladybird Harmonia axyridis. Dev Comp Immunol 49(2):303–312
Aoki W, Ueda M (2013) Characterization of antimicrobial peptides toward the development of novel antibiotics. Pharmaceuticals 6(8):1055–1081
Bogomolovas J et al (2009) Screening of fusion partners for high yield expression and purification of bioactive viscotoxins. Protein Expr Purif 64(1):16–23
Zhou L et al (2009) Expression and purification the antimicrobial peptide CM4 in Escherichia coli. Biotech Lett 31(3):437–441
Zhou L et al (2009) TrxA mediating fusion expression of antimicrobial peptide CM4 from multiple joined genes in Escherichia coli. Protein Expr Purif 64(2):225–230
Li X, Jiang Y, Lin Y (2022) Production of antimicrobial peptide arasin-like Sp in Escherichia coli via an ELP-intein self-cleavage system. J Biotechnol 347:49–55
Zorko M, Jerala R (2010) Production of recombinant antimicrobial peptides in bacteria. Antimicrobial Peptides. Springer, Berlin, pp 61–76
Abd Elhameed HA et al (2019) Purification of proteins with native terminal sequences using a Ni (II)-cleavable C-terminal hexahistidine affinity tag. Protein Expr Purif 159:53–59
Gwak WS et al (2018) Enhanced production of recombinant protein by fusion expression with Ssp DnaB mini-intein in the baculovirus expression system. Viruses 10(10):523
Xing L et al (2011) Streamlined protein expression and purification using cleavable self-aggregating tags. Microb Cell Fact 10(1):42
Gramespacher JA et al (2018) Improved protein splicing using embedded split inteins. Protein Sci 27(3):614–619
Schreiber C et al (2017) A high-throughput expression screening platform to optimize the production of antimicrobial peptides. Microb Cell Fact 16(1):1–13
Yasukawa T et al (1995) Increase of solubility of foreign proteins in escherichia coli by coproduction of the bacterial thioredoxin (∗). J Biol Chem 270(43):25328–25331
Ishida H et al (2016) Overexpression of antimicrobial, anticancer, and transmembrane peptides in Escherichia coli through a calmodulin-peptide fusion system. J Am Chem Soc 138(35):11318–11326
Supungul P et al (2008) Cloning, expression and antimicrobial activity of crustinPm1, a major isoform of crustin, from the black tiger shrimp Penaeus monodon. Dev Comp Immunol 32(1):61–70
Peng H et al (2012) Optimized production of scygonadin in Pichia pastoris and analysis of its antimicrobial and antiviral activities. Protein Expr Purif 82(1):37–44
Amparyup P et al (2008) Molecular cloning, genomic organization and recombinant expression of a crustin-like antimicrobial peptide from black tiger shrimp Penaeus monodon. Mol Immunol 45(4):1085–1093
Richard J (2017) Challenges in oral peptide delivery: lessons learnt from the clinic and future prospects. Ther Deliv 8(8):663–684
Wang C et al (2021) Antimicrobial peptides towards clinical application: Delivery and formulation. Adv Drug Deliv Rev 175:113818
Author information
Authors and Affiliations
Contributions
MK, PG, and HV: drafted the manuscript. MK: drew the figures. NF and MT: reviewed and revised the manuscript. All authors read the final version of the manuscript and approved it.
Corresponding authors
Ethics declarations
Conflict of interest
Authors declare that they have no conflict of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kordi, M., Talkhounche, P.G., Vahedi, H. et al. Heterologous Production of Antimicrobial Peptides: Notes to Consider. Protein J 43, 129–158 (2024). https://doi.org/10.1007/s10930-023-10174-w
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-023-10174-w