Abstract
Main conclusion
MYB transcription factors are essential for diverse biology processes in plants. This review has focused on the potential molecular actions of MYB transcription factors in plant immunity.
Abstract
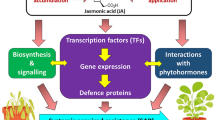
Plants possess a variety of molecules to defend against disease. Transcription factors (TFs) serve as gene connections in the regulatory networks controlling plant growth and defense against various stressors. As one of the largest TF families in plants, MYB TFs coordinate molecular players that modulate plant defense resistance. However, the molecular action of MYB TFs in plant disease resistance lacks a systematic analysis and summary. Here, we describe the structure and function of the MYB family in the plant immune response. Functional characterization revealed that MYB TFs often function either as positive or negative modulators towards different biotic stressors. Moreover, the MYB TF resistance mechanisms are diverse. The potential molecular actions of MYB TFs are being analyzed to uncover functions by controlling the expression of resistance genes, lignin/flavonoids/cuticular wax biosynthesis, polysaccharide signaling, hormone defense signaling, and the hypersensitivity response. MYB TFs have a variety of regulatory modes that fulfill pivotal roles in plant immunity. MYB TFs regulate the expression of multiple defense genes and are, therefore, important for increasing plant disease resistance and promoting agricultural production.


Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Al-Attala MN, Wang X, Abou-Attia MA, Duan X, Kang Z (2014) A novel TaMYB4 transcription factor involved in the defence response against Puccinia striiformis f. sp. tritici and abiotic stresses. Plant Mol Biol 84:589–603
Ambawat S, Sharma P, Yadav NR, Yadav RC (2013) MYB transcription factor genes as regulators for plant responses: an overview. Physiol Mol Biol Plants 19(3):307–321
An C, Sheng LP, Du XP, Wang YJ, Zhang Y, Song AP, Jiang JF, Guan ZY, Fang WM, Chen FD, Chen S (2019) Overexpression of CmMYB15 provides chrysanthemum resistance to aphids by regulating the biosynthesis of lignin. Hortic Res 6:84–94
Aoyagi LN, Lopes-Caitar VS, de Carvalho MCCG, Darben LM, Polizel-Podanosqui A, Kuwahara MK, Nepomuceno AL, Abdelnoor RV, Marcelino-Guimarães FC (2014) Genomic and transcriptomic characterization of the transcription factor family R2R3-MYB in soybean and its involvement in the resistance responses to Phakopsora pachyrhizi. Plant Sci 229:32–42
Beneteau J, Renard D, Marché L, Douville E, Lavenant L, Rahbé Y, Dupont D, Vilaine F, Dinant D (2010) Binding properties of the N-acetylglucosamine and high-mannose N-glycan PP2-A1 phloem lectin in Arabidopsis. Plant Physiol 153(3):1345–1361
Borisjuk N, Hrmova M, Lopato S (2014) Transcriptional regulation of cuticle biosynthesis. Biotechnol Adv 32:526–540
Chang C, Yu D, Jiao J, Jing S, Schulze-lefert P, Shen QH (2013) Barley MLA immune receptors directly interfere with antagonistically acting transcription factors to initiate disease resistance signaling. Plant Cell 25(3):1158–1173
Chen L, Hu B, Qin Y, Hu G, Zhao J (2019) Advance of the negative regulation of anthocyanin biosynthesis by MYB transcription factors. Plant Physiol Biochem 136:178–187
Chen M (2012) Cloning and functional analysis of rice defense-related gene OsDRxoc1 and OsDRxoc2. Dissertation, Shandong Agricultural University
ChezemWR MA, Li FS, Weng JK, Clay NK (2017) SG2-type R2R3-MYB transcription factor MYB15 controls defense-induced lignification and basal immunity in Arabidopsis. Plant Cell 29(8):1907–1926
Chi Y, Yang Y, Zhou Y, Zhou J, Fan B, Yu JQ, Chen Z (2013) Protein–protein interactions in the regulation of WRKY transcription factors. Mol Plant 6(2):287–300
Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, Leonhardt N, Dellaporta SL, Tonelli C (2005) A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol 15(13):1196–1200
Cui J, Jiang N, Zhou X, Hou X, Yang G, Meng J, Luan Y (2018) Tomato MYB49 enhances resistance to Phytophthora infestans and tolerance to water deficit and salt stress. Planta 248:1487–1503
Deng YX, Lu SF (2017) Biosynthesis and regulation of phenylpropanoids in plants. Annu Rev Plant Biol 36(4):257–290
Derksen H, Rampitsch C, Daayf F (2013) Signaling cross-talk in plant disease resistance. Plant Sci 207:79–87
Dinant S, Clark AM, Zhu Y, Vilaine F, Palauqui JC, Kusiak C, Thompson GA (2003) Diversity of the superfamily of phloem lectins (phloem protein 2) in angiosperms. Plant Physiol 131(2):114–128
Ding A, Yang X, Yu X, Chen Z, Liu Y, Wang W, Sun Y (2022) A chimeric AtERF4 repressor modulates pleiotropic aspects of plant growth and abiotic stress tolerance in transgenic Arabidopsis. Plant Growth Regul 96:255–267
Dixon RA, Pasinetti GM (2010) Flavonoids and isoflavonoids: from plant biology to agriculture and neuroscience. Plant Physiol 154(2):453–457
Du H, Wang YB, Xie Y, Liang Z, Jiang SJ, Zhang SS, Huang YB, Tang YX (2013) Genome-wide identification and evolutionary and expression analyses of MYB-related genes in land plants. DNA Res 20(5):437–448
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15(10):573–581
Ellinger D, Voigt CA (2014) Callose biosynthesis in Arabidopsis with a focus on pathogen response: what we have learned within the last decade. ANN Bot 114(6):1349–1358
Froidure S, Canonne J, Daniel X, Jauneau A, Brière C, Roby D, Rivas S (2010) AtsPLA2-α nuclear relocalization by the Arabidopsis transcription factor AtMYB30 leads to repression of the plant defense response. Proc Natl Acad Sci U S A 107(34):15281–15286
Gao Y, Jia S, Wang C, Wang F, Wang F, Zhao K (2016) BjMYB1, a transcription factor implicated in plant defence through activating BjCHI1 chitinase expression by binding to a W-box-like element. J Exp Bot 67(15):4647–4658
Gao Y, Zan XL, Wu XF, Yao L, Chen YL, Jia SW, Zhao KJ (2014) Identification of fungus-responsive cis-acting element in the promoter of Brassica juncea chitinase gene, BjCHI1. Plant Sci 215–216:190–198
Geng P, Zhang S, Liu J, Zhao C, Wu J, Cao Y, Fu C, Han X, He H, Zhao Q (2020) MYB20, MYB42, MYB43, and MYB85 regulate phenylalanine and lignin biosynthesis during secondary cell wall formation. Plant Physiol 182(3):1272–1283
Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Ann Rev Phytopathol 43:205–227
Gu KD, Zhang QY, Yu JQ, Wang JH, Zhang FJ, Wang CK, Zhao YW, Sun CH, You CX, Hu DG, Hao YJ (2020) R2R3-MYB transcription factor MdMYB73 confers increased resistance to the fungal pathogen Botryosphaeria dothidea in apples via the salicylic acid pathway. J Agric Food Chem 69(1):447–458
Gu Z, Zhu J, Zhang L, Yang Y, Li S, Wang L (2019) Transcription factors participate in response to powdery mildew infection in Paeonia lactiflora. Sci Hortic 17:108535
Haga N, Kato K, Murase M, Araki S, Kubo M, Demura T, Suzuki K, Müller I, Voß U, Jürgens G, Ito M (2007) R1R2R3-Myb proteins positively regulate cytokinesis through activation of KNOLLE transcription in Arabidopsis thaliana. Development 134(6):1101–1110
Hayashi K, Fujita Y, Ashizawa T, Suzuki F, Nagamura Y, Hayano-Saito Y (2016) Serotonin attenuates biotic stress and leads to lesion browning caused by a hypersensitive response to Magnaporthe oryzae penetration in rice. Plant J 85(1):46–56
He J, Liu Y, Yuan D, Duan M, Liu Y, Shen Z, Yang C, Qiu Z, Li D, Wen P, Huang J, Fan D, Xiao S, Xin Y, Chen X, Jiang L, Wang H, Yuan L, Wan J (2020) An R2R3 MYB transcription factor confers brown planthopper resistance by regulating the phenylalanine ammonia-lyase pathway in rice. Proc Natl Acad Sci U S A 117(1):271–277
Hoseinzadeh AH, Soorni A, Shoorooei M, Mahani MT, Amiri RM, Allahyari H, Mohammadi R (2020) Comparative transcriptome provides molecular insight into defense-associated mechanisms against spider mite in resistant and susceptible common bean cultivars. PLoS ONE 15(2):e0228680
Hu Q, Min L, Yang X, Jin S, Zhang L, Li Y, Ma Y, Qi X, Li D, Liu H, Lindsey K, Zhu L, Zhang X (2018) Laccase GhLac1 modulates broad-spectrum biotic stress tolerance via manipulating phenylpropanoid pathway and jasmonic acid synthesis. Plant Physiol 176:1808–1823
Hu G (2018) miR319-MYB33-SPL9-DFR pathway participates upland cotton response against Verticillium dahliae. Dissertation, Jishou University
Jia L, Clegg MT, Jiang T (2004) Evolutionary dynamics of the DNA-binding domains in putative R2R3-MYB genes identified from rice subspecies indica and japonica genomes. Plant Physiol 134(2):575–585
Jia N, Liu J, Sun Y, Tan P, Cao H, Xie Y, Wen B, Gu T, Liu J, Li M, Huang Y, Lu J, Jin N, Sun L, Xin F, Fan B (2018) Citrus sinensis MYB transcription factors CsMYB330 and CsMYB308 regulate fruit juice sac lignification through fine tuning expression of the Cs4CL1 gene. Plant Sci 277:334–343
Jia J, Xing J, Dong J, Han J, Liu J (2011) Function analysis of MYB73 of Arabidopsis thaliana against Bipolaris oryzae. Agri Sci in China 10(5):721–727
Jiang JY, Liao XL, Jin XY, Tan L, Lu QF, Yuan CL, Xue YF, Yin NW, Lin N, Chai YR (2020) MYB43 in oilseed rape (Brassica napus) positively regulates vascular lignification, plant morphology and yield potential but negatively affects resistance to sclerotinia sclerotiorum. Genes (basel) 11(5):581–604
Jiang C (2021) Mechanism analysis of transcription factor VqMYB154 regulating the resistance to powdery mildew in Chinese wild Vitis quinquangularis. Dissertation, Northwest Agriculture and Forestry University
Katiyar A, Smita S, Lenka SK, Rajwanshi R, Chinnusamy V, Bansal KC (2012) Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genom 13:544
Kelemen Z, Sebastian A, Xu W, Grain D, Salsac F, Avon A, Berger N, Tran J, Dubreucq B, Lurin C, Lepiniec L, Contreras-Moreira B, Dubos C (2015) Analysis of the DNA-binding activities of the Arabidopsis R2R3-MYB transcription factor family by one-hybrid experiments in yeast. PLoS ONE 10(10):e141044
Kim SH, Lam PY, Lee MH, Jeon HS, Tobimatsu Y, Park OK (2020) The Arabidopsis R2R3 MYB transcription factor MYB15 is a key regulator of lignin biosynthesis in effector-triggered immunity. Front Plant Sci 11:583153–583163
Kishi-Kaboshi M, Seo S, Takahashi A, Hirochika H (2018) The MAMP-responsive MYB transcription factors MYB30, MYB55 and MYB110 activate the HCAA synthesis pathway and enhance immunity in rice. Plant Cell Physiol 59(5):903–915
Klempnauer KH, Gonda TJ, Michael Bishop J (1982) Nucleotide sequence of the retroviral leukemia gene v-myb and its cellular progenitor c-myb: the architecture of a transduced oncogene. Cell 31:453–463
Lee MW, Qi M, Yang Y (2001) A novel jasmonic acid-inducible rice myb gene associates with fungal infection and host cell death. Mol Plant Microbe Interact 14(4):527–535
Li J, Han G, Sun C, Sui N (2019) Research advances of MYB transcription factors in plant stress resistance and breeding. Plant Signal Behav 14(8):1613131
Li C, Ng CKY, Fan LM (2015) MYB transcription factors, active players in abiotic stress signaling. Environ Exp Bot 114:80–91
Li W, Wang K, Chern M, Chern M, Liu Y, Zhu Z, Liu J, Zhu X, Yin J, Ran L, Xiong J, He K, Xu L, He M, Wang J, Liu J, Bi Y, Qing H, Li M, Hu K, Song L, Wang L, Qi T, Hou Q, Chen W, Li Y, Wang W, Chen X (2020) Sclerenchyma cell thickening through enhanced lignification induced by OsMYB30 prevents fungal penetration of rice leaves. New Phytol 226(6):1850–1863
Li T, Zhang XY, Huang Y, Xu ZS, Wang F, Xiong AS (2018) An R2R3-MYB transcription factor, SlMYB28, involved in the regulation of TYLCV infection in tomato. Sci Hortic 237:192–200
Li Q, Zhang C, Li J, Wang L, Ren Z (2012) Genome-wide identification and characterization of R2R3MYB family in Cucumis sativus. PLoS ONE 7(10):e47576
Li M (2012) The heterologous expression of flavonoid specific regulatory factor AtMYB12 gene and the contributions to plant resistance. Dissertation, Shandong Agricultural University
Liu R, Chen L, Jia Z, Lü B, Shi H, Shao W, Dong H (2011) Transcription factor AtMYB44 regulates induced expression of the ETHYLENE INSENSITIVE2 gene in Arabidopsis responding to a harpin protein. Mol Plant Microbe Interact 24(3):377–389
Liu T, Chen T, Kan J, Yao Y, Guo D, Yang Y, Ling X, Wang J, Zhang B (2022) The GhMYB36 transcription factor confers resistance to biotic and abiotic stress by enhancing PR1 gene expression in plants. Plant Biotechnol J 20(4):722–735
Liu Y, Li M, Li T, Chen Y, Zhang L, Zhao G, Zhuang J, Zhao W, Gao L, Xia T (2020) Airborne fungus-induced biosynthesis of anthocyanins in Arabidopsis thaliana via jasmonic acid and salicylic acid signaling. Plant Sci 300:110635
Liu Z, Luan Y, Li J, Yin Y (2016) Expression of a tomato MYB gene in transgenic tobacco increases resistance to Fusarium oxysporum and Botrytis cinerea. Eur J Plant Pathol 144:607–617
Liu R, Lü B, Wang X, Zhang C, Zhang S, Qian J, Chen L, Shi H, Dong H (2010) Thirty-seven transcription factor genes differentially respond to a harpin protein and affect resistance to the green peach aphid in Arabidopsis. J Biosci 35:435–450
Liu J, Osbourn A, Ma P (2015) MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol Plant 8:689–708
Liu X, Yang L, Zhou X, Zhou M, Lu Y, Ma L, Ma H, Zhang Z (2013) Transgenic wheat expressing Thinopyrum intermedium MYB transcription factor TiMYB2R-1 shows enhanced resistance to the take-all disease. J Exp Bot 64(8):2243–2253
Lokesh U, Venkatesh B, Kiranmai K, Kumar AN, Reddy VA, Kirankumar TV, Sudhakar C (2016) Transcriptional regulation of functional genes involved in cuticular wax biosynthesis by MYB family transcriptional factors in plants. International Journal of Environmental and Agriculture Research 2(3):86–92
Luo L (2018) The transcriptional regulatory mechanism of the poplar transcription factor MYB6 involved in flavonoid and lignin biosynthesis. Dissertation, Southwest University
Lyu Q (2022) Wheat MYB28 regulates wax biosynthesis and resistance to insect and pathogen attacks. Dissertation, Shandong Agricultural University
Lü BB, Li XJ, Sun WW, Li L, Gao R, Zhu Q, Tian SM, Fu MQ, Yu HL, Tang XM, Zhang CL, Dong HS (2013) AtMYB44 regulates resistance to the green peach aphid and diamondback moth by activating EIN2-affected defences in Arabidopsis. Plant Biol 15(5):841–850
Ma R, Liu B, Geng X, Ding X, Yan N, Sun X, Wang W, Sun X, Zheng C (2023) Biological function and stress response mechanism of MYB transcription factor family genes. J Plant Growth Regul 42:83–95
Mabuchi K, Maki H, Itaya T, Suzuki T, Nomoto M, Sakaoka S, Morikami A, Higashiyama T, Tada Y, Busch W, Tsukagoshi H (2018) MYB30 links ROS signaling, root cell elongation, and plant immune responses. Proc Natl Acad Sci U S A 115(20):E4710–E4719
Matus JT, Aquea F, Arce-Johnson P (2008) Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biol 8:83
Meng X, Yu Y, Song T, Yu Y, Cui N, Ma Z, Chen L, Fan H (2022) Transcriptome sequence analysis of the defense responses of resistant and susceptible cucumber strains to Podosphaera xanthii. Front Plant Sci 13:872218
Mengiste T, Chen X, Salmeron J, Dietrich R (2003) The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 15(11):2551–2565
Ngou B, Ahn HK, Ding P, Jones J (2021) Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592(7852):110–115
Noman A, Hussain A, Adnan M, Khan MI, Ashraf MF, Zainab M, Khan KA, Ghramh HA, He S (2019) A novel MYB transcription factor CaPHL8 provide clues about evolution of pepper immunity against soil borne pathogen. Microb Pathog 137:103758
Ogata K, Kanei-Ishi C, Sasaki M, Hatanaka H, Nagadoi A, Enari M, Nakamura H, Nishimura Y, Ishii S, Sarai A (1996) The cavity in the hydrophobic core of Myb DNA-binding domain is reserved for DNA recognition and trans-activation. Nat Struct Biol 3(2):178–187
Ohtani M, Demura T (2019) The quest for transcriptional hubs of lignin biosynthesis: beyond the NAC-MYB-gene regulatory network model. Curr Opin Biotechnol 56:82–87
Paz-Ares J, Ghosal D, Wienand U, Peterson PA, Saedler H (1987) The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J 6:3553–3558
Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W, Ren C, Xie D (2011) The jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23(5):1795–1814
Qiang Z, Sun H, Ge F, Li W, Li C, Wang S, Zhang B, Zhu L, Zhang S, Wang X, Lai J, Qin F, Zhou Y, Fu Y (2022) The transcription factor ZmMYB69 represses lignin biosynthesis by activating ZmMYB31/42 expression in maize. Plant Physiol 189(4):1916–1919
Qiu Z, Yan S, Xia B, Jiang J, Yu B, Lei J, Chen C, Chen L, Yang Y, Wang Y, Tian S, Cao B (2019) The eggplant transcription factor MYB44 enhances resistance to bacterial wilt by activating the expression of spermidine synthase. J Exp Bot 70(19):5343–5354
Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W (2003) Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol 133(3):1051–1071
Raffaele S, Rivas S (2013) Regulate and be regulated: integration of defense and other signals by the AtMYB30 transcription factor. Front Plant Sci 4:98
Raffaele S, Vailleau F, Léger A, Joubès J, Miersch O, Huard C, Blée E, Mongrand S, Domergue F, Roby D (2008) A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 20(3):752–767
Ramsay NA, Glover BJ (2005) MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci 10(2):63–70
Romero I, Fuertes A, Benito MJ, Malpica JM, Leyva A, Paz-Ares J (1998) More than 80R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana. Plant J 14(3):273–284
Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15(5):247–258
Salih H, Gong W, He S, Sun G, Sun J, Du X (2016) Genome-wide characterization and expression analysis of MYB transcription factors in Gossypium hirsutum. BMC Genet 17(1):129
Schaffer R, Landgraf J, Accerbi M, Silmon VA, Larson M, Wisman E (2001) Microarray analysis of diurnal and circadian- regulated genes in Arabidopsis. Plant Cell 13(1):113–123
Seo PJ, Park CM (2010) MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. New Phytol 186:471–483
Shan T, Rong W, Xu H, Du L, Liu X, Zhang Z (2016) The wheat R2R3-MYB transcription factor TaRIM1 participates in resistance response against the pathogen Rhizoctonia cerealis infection through regulating defense genes. Sci Rep 6:28777
Shen X, Guo X, Guo X, Zhao D, Zhao W, Chen J, Li T (2017) PacMYBA, a sweet cherry R2R3-MYB transcription factor, is a positive regulator of salt stress tolerance and pathogen resistance. Plant Physiol Biochem 112:302–311
Shen JL (2020) Analysis of the mechanism of GhMYB43 regulating cotton resistance to Verticillium dahliae. Dissertation, Shihezi University
Shi H (2010) Function analysis of transcription factor AtMYB44 and AtMYB15 during defense response in Arabidopsis. Dissertation, Nanjing Agricultural University
Spoel SH, Johnson JS, Dong X (2007) Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc Natl Acad Sci U S A 104(47):18842–18847
Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4(5):447–456
Treutter D (2008) Significance of flavonoids in plant resistance and enhancement of their biosynthesis. Plant Biol 7(6):581–590
Vailleau F, Daniel X, Tronchet M, Montillet JL, Triantaphylidès C, Roby D (2002) A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc Natl Acad Sci U S A 99(15):10179–10184
Wang Y, Liu W, Jiang H, Mao Z, Wang N, Jiang S, Xu H, Yang G, Zhang Z, Chen X (2019) The R2R3-MYB transcription factor MdMYB24-like is involved in methyl jasmonate-induced anthocyanin biosynthesis in apple. Plant Physiol Biochem 139:273–282
Wang Y, Sheng L, Zhang H, Du X, An C, Xia X, Chen F, Jiang J, Chen S (2017) CmMYB19 over-expression improves aphid tolerance in chrysanthemum by promoting lignin synthesis. Int J Mol Sci 18(3):619–632
Wang G (2016) Molecular mechanism of transcription factor AtMYB73 against Pst DC3000 and Botrytis cinerea in Arabidopsis thaliana. Dissertation, Hebei Agricultural University
Wei Q, Chen R, Wei X, Liu Y, Zhao S, Yin X, Xie T (2020) Genome-wide identification of R2R3-MYB family in wheat and functional characteristics of the abiotic stress responsive gene TaMYB344. BMC Genom 21(1):792
Wilkins O, Nahal H, Foong J, Provart NJ, Campbell MM (2009) Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol 149(2):981–993
Will T, van Bel AJ (2006) Physical and chemical interactions between aphids and plants. J Exp Bot 57(4):729–737
Wu W (2020) The molecular mechanism analysis of OsDRxoc1-mediated resistance to bacterial leaf streak. Dissertation, Shandong Agricultural University
Xiao S, Hu Q, Shen J, Liu S, Yang Z, Chen K, Klosterman SJ, Javornik B, Zhang X, Zhu L (2021) GhMYB4 downregulates lignin biosynthesis and enhances cotton resistance to Verticillium dahliae. Plant Cell Rep 40:735–751
Xu J (2009) Study on the inhibitory effect of AtMYB13 on defense response of Arabidopsis thaliana. Dissertation, Nanjing Agricultural University
Yao L, Yang B, Xian B, Chen B, Yan J, Chen Q, Gao S, Zhao P, Han F, Xu J, Jiang Y (2020) The R2R3-MYB transcription factor BnaMYB111L from rapeseed modulates reactive oxygen species accumulation and hypersensitive-like cell death. Plant Physiol Biochem 147:280–288
Yu EY, Kim SE, Kim JH, Ko JH, Cho MH, Chung IK (2000) Sequence-specific DNA recognition by the Myb-like domain of plant telomeric protein RTBP1. J Biol Chem 275(31):24208–24214
Zhai Y, Li P, Mei Y, Chen M, Chen X, Xu H, Zhou X, Dong H, Zhang C, Jiang W (2017) Three MYB genes co-regulate the phloem-based defence against English grain aphid in wheat. J Exp Bot 68(15):4153–4169
Zhang Z, Liu X, Wang X, Zhou M, Zhou X, Ye X, Wei X (2012) An R2R3 MYB transcription factor in wheat, TaPIMP1, mediates host resistance to Bipolaris sorokiniana and drought stresses through regulation of defense- and stress-related genes. New Phytol 196(4):1155–1170
Zhang B, Schader A (2017) TRANSPARENT TESTA GLABRA 1-dependent regulation of flavonoid biosynthesis. Plants 6(4):5
Zhang CL, Shi HJ, Chen L, Wang X, Lü B, Zhang S, Liang Y, Liu R, Qian J, Sun W, You ZZ, Dong H (2011) Harpin-induced expression and transgenic overexpression of the phloem protein gene AtPP2-A1 in Arabidopsis repress phloem feeding of the green peach aphid Myzus persicae. BMC Plant Biol 11:11
Zhang M, Wang J, Luo Q, Yang C, Yang H, Cheng Y (2021) CsMYB96 enhances citrus fruit resistance against fungal pathogen by activating salicylic acid biosynthesis and facilitating defense metabolite accumulation. J Plant Physiol 264:135472
Zhang YL, Zhang CL, Wang GL, Wang YX, Qi CH, Zhao Q, You CX, Li YY, Hao YJ (2019) The R2R3MYB transcription factor MdMYB30 modulates plant resistance against pathogens by regulating cuticular wax biosynthesis. BMC Plant Biol 19(1):362
Zhao P, Li Q, Li J, Wang L, Ren Z (2014) Genome-wide identification and characterization of R2R3MYB family in Solanum lycopersicum. Mol Genet Genomics 289(6):1183–1207
Zheng H, Dong L, Han X, Jin H, Yin C, Han Y, Li B, Qin H, Zhang J, Shen Q, Zhang K, Wang D (2020) The TuMYB46L-TuACO3 module regulates ethylene biosynthesis in einkorn wheat defense to powdery mildew. New Phytol 225(6):2526–2541
Zhu Y, Hu X, Wang P, Wang H, Ge X, Li F, Hou Y (2022) GhODO1, an R2R3-type MYB transcription factor, positively regulates cotton resistance to Verticillium dahliae via the lignin biosynthesis and jasmonic acid signaling pathway. Int J Biol Macromol 201:580–591
Acknowledgements
This study was funded by National Natural Science Foundation of China (32102366), Natural Science Foundation of Liaoning Province of China (2020-BS-138) and Project of Education Department of Liaoning Province (LJKMZ20221809).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Additional information
Communicated by Gerhard Leubner.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yu, Y., Zhang, S., Yu, Y. et al. The pivotal role of MYB transcription factors in plant disease resistance. Planta 258, 16 (2023). https://doi.org/10.1007/s00425-023-04180-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-023-04180-6