Abstract
Main conclusion
Opposing changes in the abundance of satellite DNA and long terminal repeat (LTR) retroelements are the main contributors to the variation in genome size and heterochromatin amount in Arachis diploids.
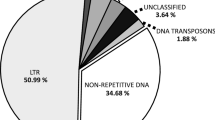
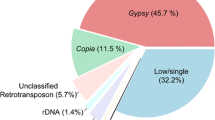
The South American genus Arachis (Fabaceae) comprises 83 species organized in nine taxonomic sections. Among them, section Arachis is characterized by species with a wide genome and karyotype diversity. Such diversity is determined mainly by the amount and composition of repetitive DNA. Here we performed computational analysis on low coverage genome sequencing to infer the dynamics of changes in major repeat families that led to the differentiation of genomes in diploid species (x = 10) of genus Arachis, focusing on section Arachis. Estimated repeat content ranged from 62.50 to 71.68% of the genomes. Species with different genome composition tended to have different landscapes of repeated sequences. Athila family retrotransposons were the most abundant and variable lineage among Arachis repeatomes, with peaks of transpositional activity inferred at different times in the evolution of the species. Satellite DNAs (satDNAs) were less abundant, but differentially represented among species. High rates of evolution of an AT-rich superfamily of satDNAs led to the differential accumulation of heterochromatin in Arachis genomes. The relationship between genome size variation and the repetitive content is complex. However, largest genomes presented a higher accumulation of LTR elements and lower contents of satDNAs. In contrast, species with lowest genome sizes tended to accumulate satDNAs in detriment of LTR elements. Phylogenetic analysis based on repetitive DNA supported the genome arrangement of section Arachis. Altogether, our results provide the most comprehensive picture on the repeatome dynamics that led to the genome differentiation of Arachis species.






Similar content being viewed by others
Abbreviations
- DAPI:
-
4′,6-Diamidino-2-phenylindole
- FISH:
-
Fluorescent in situ hybridization
- GISH:
-
Genome in situ hybridization
- LTR:
-
Long terminal repeat
- MYA:
-
Million years ago
- rDNA:
-
Ribosomal DNA
- satDNA:
-
Satellite DNA
- RepeatExplorer2:
-
RE2
- MULEs:
-
Mutator-like elements
References
Alioto T, Alexiou KG, Bardil A et al (2020) Transposons played a major role in the diversification between the closely related almond and peach genomes: results from the almond genome sequence. Plant J 101:455–472
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Ambrožová K, Mandáková T, Bureš P, Neumann P, Leitch IJ, Koblížková A, Macas J, Lysak MA (2011) Diverse retrotransposon families and an AT-rich satellite DNA revealed in giant genomes of Fritillaria lilies. Ann Bot 107:255–268
Ammiraju JS, Zuccolo A, Yu Y, Song X, Piegu B, Chevalier F, Walling JG, Ma J, Talag J, Brar DS, SanMiguel PJ, Jiang N, Jackson SA, Panaud O, Wing RA (2007) Evolutionary dynamics of an ancient retrotransposon family provides insights into evolution of genome size in the genus Oryza. Plant J 52:342–351
Bechara MD, Moretzsohn MC, Palmieri DA, Monteiro JP, Bacci M, Martins J, Valls JFM, Lopes CR, Gimenes MA (2010) Phylogenetic relationships in genus Arachis based on ITS and 5.8 S rDNA sequences. BMC Plant Biol 10:255
Bennett MD (1987) Variation in genomic form in plants and its ecological implications. New Phytol 106:177–200
Bennetzen JL (2005) Transposable elements, gene creation and genome rearrangement in flowering plants. Curr Opin Genet Dev 15:621–627
Bennetzen JL, Kellogg EA (1997) Do plants have a one-way ticket to genomic obesity? Plant Cell 9:1509–1514
Bennetzen JL, Wang H (2014) The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annu Rev Plant Biol 65:505–530
Bertioli DJ, Vidigal B, Nielen S, Ratnaparkhe MB, Lee TH, Leal-Bertioli SCM, Kim C, Guimarães PM, Seijo G, Schwarzacher T, Paterson AH, Heslop-Harrison P, Araujo ACG (2013) The repetitive component of the A genome of peanut (Arachis hypogaea) and its role in remodelling intergenic sequence space since its evolutionary divergence from the B genome. Ann Bot 112:545–559
Bertioli DJ, Cannon SB, Froenicke L et al (2016) The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat Genet 8:438–446
Bertioli DJ, Jenkins J, Clevenger J et al (2019) The genome of segmental allotetraploid peanut Arachis hypogaea. Nat Genet 51:877–884
Biémont C, Vieira C (2006) Junk DNA as an evolutionary force. Nature 443:521–524
Biscotti MA, Olmo E, Heslop-Harrison JP (2015) Repetitive DNA in eukaryotic genomes. Chromosome Res 23:415–420
Bolsheva NL, Melnikova NV, Kirov IV, Dmitriev AA, Krasnov GS, Amosova AV, Samatadze TE, Yurkevich OY, Zoshchuk SA, Kudryavtseva AV, Muravenko OV (2019) Characterization of repeated DNA sequences in genomes of blue-flowered flax. BMC Evol Biol 19:49
Chenais B, Caruso A, Hiard S, Casse N (2012) The impact of transposable elements on eukaryotic genomes: from genome size increase to genetic adaptation to stressful environments. Gene 509:7–15
Darzentas N (2010) Circoletto: visualizing sequence similarity with Circos. Bioinformatics 26:2620–2621
Devos KM, Brown JKM, Bennetzen JL (2002) Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res 12:1075–1079
Dhillon SS, Rake AV, Miksche JP (1980) Reassociation kinetics and cytophotometric characterisation of peanut (Arachis hypogaea L.) DNA. Plant Physiol 65:1121–1127
Di Rienzo JA, Casanoves F, Balzarini MG, González L, Tablada M, Robledo CW (2013) InfoStat version 2013. Grupo InfoStat, FCA, Universidad Nacional de Córdoba, Argentina. http://www.infostat.com.ar
Dodsworth S, Chase MW, Kelly LJ, Leitch IJ, Macas J, Novák P, Piednoël M, Weiss-Schneeweiss H, Leitch AR (2014) Genomic repeat abundances contain phylogenetic signal. Syst Biol 64:112–126
Dodsworth S, Chase MW, Särkinen T, Knapp S, Leitch AR (2016) Using genomic repeats for phylogenomics: a case study in wild tomatoes (Solanum section Lycopersicon: Solanaceae). Biol J Linn Soc 117:96–105
Dodsworth S, Jang TS, Struebig M, Chase MW, Weiss-Schneeweiss H, Leitch AR (2017) Genome-wide repeat dynamics reflect phylogenetic distance in closely related allotetraploid Nicotiana (Solanaceae). Plant Syst Evol 303:1013–1020
Dogan M, Pouch M, Mandáková T, Hloušková P, Guo X, Winter P, Chumová Z, Van Niekerk A, Mummenhoff K, Al-Shehbaz IA, Mucina L, Lysak MA (2021) Evolution of tandem repeats is mirroring post-polyploid cladogenesis in Heliophila (Brassicaceae). Front Plant Sci 11:1944
Doležel J, Bartos J (2005) Plant DNA flow cytometry and estimation of nuclear genome size. Ann Bot 95:99–110
El Baidouri M, Panaud O (2013) Comparative genomic paleontology across plant kingdom reveals the dynamics of TE-driven genome evolution. Genome Biol Evol 5:954–965
Emadzade K, Jang TS, Macas J, Kovařík A, Novák P, Parker J, Weiss-Schneeweiss H (2014) Differential amplification of satellite PaB6 in chromosomally hypervariable Prospero autumnale complex (Hyacinthaceae). Ann Bot 114:1597–1608
Ewing AD (2015) Transposable element detection from whole genome sequence data. Mob DNA 6:24
Fernandez A, Krapovickas A (1994) Cromosomas y evolucion en Arachis (Leguminosae). Bonplandia 8:187–220
Ferree PM, Barbash DA (2009) Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila. PLoS Biol 7:e1000234
Feschotte C, Jiang N, Wessler SR (2002) Plant transposable elements: where genetics meets genomics. Nat Rev Genet 3:329–341
Fransz P, Linc G, Lee CR, Aflitos SA et al (2016) Molecular, genetic and evolutionary analysis of a paracentric inversion in Arabidopsis thaliana. Plant J 88:159–178
Friend SA, Quandt D, Tallury SP, Stalker HT, HiluK W (2010) Species, genomes, and section relationships in the genus Arachis (Fabaceae): a molecular phylogeny. Plant Syst Evol 290:185–199
Gaiero P, Vaio M, Peters SA, Schranz ME, de Jong H, Speranza PR (2019) Comparative analysis of repetitive sequences among species from the potato and the tomato clades. Ann Bot 123:521–532
Garrido-Ramos MA (2015) Satellite DNA in plants: more than just rubbish. Cytogenet Genome Res 146:153–170
Goloboff PA, Farris JS, Nixon K (2008) TNT, a free program for phylogenetic analysis. Cladistics 24:774–786
Gowda MVC, Bhat RS, Sujay V, Kusuma P, Bhat S, Varshney RK (2011) Characterization of AhMITE1 transposition and its association with the mutational and evolutionary origin of botanical types in peanut (Arachis spp.). Plant Syst Evol 291:153–158
Grabiele M, Chalup L, Robledo G, Seijo G (2012) Genetic and geographic origin of domesticated peanut as evidenced by 5S rDNA and chloroplast DNA sequences. Plant Syst Evol 298:1151–1165
Gray YH (2000) It takes two transposons to tango. Trends Genet 16:461–468
Gregory MP, Gregory WC (1979) Exotic germoplasm of Arachis L. interspecific hybrids. J Hered 70:185–193
Greilhuber J, Lysák M, Doležel J, Bennett MD (2005) The origin, evolution and proposed stabilization of the terms ‘genome size’, and ‘C-value’ to describe nuclear DNA contents. Ann Bot 95:255–260
Holland B, Moulton V (2003) Consensus networks: a method for visualising incompatibilities in collections of trees. In: Benson G, Page RDM (eds) International workshop on algorithms in bioinformatics. Springer, Berlin, pp 165–176
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267
Jurka J, Bao W, Kojima KK (2011) Families of transposable elements, population structure and the origin of species. Biol Direct 6:44
Jurka J, Bao W, Kojima KK, Kohany O, Yurka MG (2012) Distinct groups of repetitive families preserved in mammals correspond to different periods of regulatory innovations in vertebrates. Biol Direct 7:36
Katoh K, Misasa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucl Acids Res 30:3059–3066
Kent WJ (2002) BLAT—the BLAST-like alignment tool. Genome Res 12:656–664
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Krapovickas A, Gregory WC (1994) Taxonomy of the genus Arachis (Leguminosae). Bonplandia 8:1–186
Kreplak J, Madoui MA, Cápal P et al (2019) A reference genome for pea provides insight into legume genome evolution. Nat Genet 51:1411–1422
Kumar A, Bennetzen JL (1999) Plant retrotransposons. Annu Rev Genet 33:479–532
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Lavin M, Schrire BD, Lewis GP, Pennington RT, Delgado-Salinas A, Thulin M, Hughes CE, Beyra-Matos A, Wojciechowski MF (2004) Metacommunity processes rather than continental tectonic history better explain geographically structured phylogenies in legumes. Philos Trans R Soc Lond Biol Sci 359:1509–1522
Leal-Bertioli SCM, Santos SP, Dantas KM, Inglis PW, Nielen S, Araujo ACG, Silva JP, Cavalcante U, Guimarães PM, Brasileiro ACM, Carrasquilla-Garcia N, Penmetsa RV, Cook D, Moretzsohn MC, Bertioli DJ (2015) Arachis batizocoi: a study of its relationship to cultivated peanut (A. hypogaea) and its potential for introgression of wild genes into the peanut crop using induced allotetraploids. Ann Bot 115:237–249
Li W, Zhang P, Fellers JP, Friebe B, Gill BS (2004) Sequence composition, organization, and evolution of the core Triticeae genome. Plant J 40:500–511
López-Flores I, Garrido-Ramos MA (2012) The repetitive DNA content of eukaryotic genomes. Genome Dyn 7:1–28
Ma J, Bennetzen JL (2004) Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci USA 101:12404–12410
Macas J, Novák P, Pellicer J, CíŽková J, KoblíŽková A, Neumann P, Fuková I, Doležel J, Kelly LJ, Leitch IJ (2015) In depth characterization of repetitive DNA in 23 plant genomes reveals sources of genome size variation in the legume tribe fabeae. PLoS ONE 10:e0143424
Mascagni F, Usai G, Natali L, Cavallini A, Giordani T (2018) A comparison of methods for LTR-retrotransposon insertion time profiling in the Populus trichocarpa genome. Caryologia 71:85–92
Mascagni F, Vangelisti A, Giordani T, Cavallini A, Natali L (2020) A computational comparative study of the repetitive DNA in the genus Quercus L. Tree Genet Genomes 16:1–11
McCann J, Macas J, Novák P, Stuessy TF, Villaseñor JL, Weiss-Schneeweiss H (2020) Differential genome size and repetitive DNA evolution in diploid species of Melampodium sect. Melampodium (Asteraceae). Front Plant Sci 11:362
Mehrotra S, Goyal V (2014) Repetitive sequences in plant nuclear DNA: types, distribution, evolution and function. Genom Proteom Bioinform 12:164–171
Moretzsohn MC, Hopkins MS, Mitchell SE, Kresovich S, Valls JFM, Ferreira ME (2004) Genetic diversity of peanut (Arachis hypogaea L.) and its wild relatives based on the analysis of hypervariable regions of the genome. BMC Plant Biol 4:11
Moretzsohn MC, Gouvea EG, Inglis PW, Leal-Bertioli SCM, Valls JFM, Bertioli DJ (2013) A study of the relationships of cultivated peanut (Arachis hypogaea) and its most closely related wild species using intron sequences and microsatellite markers. Ann Bot 111:113–126
Negm S, Greenberg A, Larracuente AM, Sproul JS (2021) RepeatProfiler: a pipeline for visualization and comparative analysis of repetitive DNA profiles. Mol Ecol Resour 21:969–981
Nelson MG, Linheiro RS, Bergman CM (2017) McClintock: an integrated pipeline for detecting transposable element insertions in whole-genome shotgun sequencing data. G3: Genes Genom Genet 7:2763–2778
Nielen S, Campos-Fonseca F, Leal-Bertioli SCM, Guimaraes PM, Seijo G, Town C, Arrial R, Bertioli D (2010) FIDEL—a retrovirus-like retrotransposon and its distinct evolutionary histories in the A and B-genome components of cultivated peanut. Chromosome Res 18:227–246
Nielen S, Vidigal B, Leal-Bertioli SCM, Ratnaparkhe M, Paterson A, Garsmeur O, D’Hont A, Guimaraes P, Bertioli D (2012) Matita, new retroelement from peanut: characterization and evolutionary context in the light of the Arachis A–B genome divergence. Mol Genet Genomics 287:21–38
Noé L, Kucherov G (2005) YASS: enhancing the sensitivity of DNA similarity search. Nucl AcIds Res 33:540–543
Novák P, Robledillo LA, Koblızková A, Vrbová I, Neumann P, Macas J (2017) TAREAN: a computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucl Acids Res 45:e111
Novák P, Neumann P, Macas J (2020) Global analysis of repetitive DNA from unassembled sequence reads using repeatexplorer2. Nat Protoc 15:3745–3776
Ortiz AM, Robledo G, Seijo G, Valls JFM, Lavia GI (2017) Cytogenetic evidences on the evolutionary relationships between the tetraploids of the section Rhizomatosae and related diploid species (Arachis, Leguminosae). J Plant Res 130:791–807
Paradis E, Schliep K (2018) Ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35:526–528
Patel M, Jung S, Moore K, Powell G, Ainsworth C, Abbott A (2004) High-oleate peanut mutants result from a MITE insertion into the FAD2 gene. Theor Appl Genet 108:1492–1502
Piegu B, Guyot R, Picault N, Roulin A, Sanyal A, Kim H, Collura K, Brar DS, Jackson S, Wing RA, Panaud O (2006) Doubling genome size without polyploidization: dynamics of retrotransposition driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res 16:1262–1269
Plohl M, Luchetti A, Mestrovic N, Mantovani B (2008) Satellite DNAs between selfishness and functionality: structure, genomics and evolution of tandem repeats in centromeric (hetero) chromatin. Gene 409:72–82
Plohl M, Meštrović N, Mravinac B (2012) Satellite DNA evolution. Genome Dyn 7:126–152
Plohl M, Meštrović N, Mravinac B (2014) Centromere identity from the DNA point of view. Chromosoma 123:313–325
R Core Team (2018) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Ren L, Huang W, Cannon EKS, Bertioli DJ, Cannon SB (2018) A mechanism for genome size reduction following genomic rearrangements. Front Genet 9:454–454
Ribeiro T, Vasconcelos E, Dos Santos KGB, Vaio M, Brasileiro-Vidal AC, Pedrosa-Harand A (2020) Diversity of repetitive sequences within compact genomes of Phaseolus L. beans and allied genera Cajanus L. and Vigna Savi. Chromosome Res 28:139–153
Robledillo LA, Koblížkova A, Petr N, Böttinger K, Vrbová I, Neumann P, Schubert I, Macas J (2018) Satellite DNA in Vicia faba is characterized by remarkable diversity in its sequence composition, association with centromeres, and replication timing. Sci Rep 8:5838
Robledo G, Seijo JG (2008) Characterization of Arachis D genome using physical mapping of heterochromatic regions and rDNA loci by FISH. Genet Mol Biol 31:717–724
Robledo G, Seijo G (2010) Species relationships among the wild B genome of Arachis species (section Arachis) based on FISH mapping of rDNA loci and heterochromatin detection: a new proposal for genome arrangement. Theor App Genet 121:1033–1046
Robledo G, Lavia GI, Seijo G (2009) Species relations among wild Arachis species with the A genome as revealed by FISH mapping of rDNA loci and heterochromatin detection. Theor App Genet 118:1295–1307
Sader M, Vaio M, Cauz-Santos LA, Dornelas MC, Vieira MLC, Melo N, Pedrosa-Harand A (2021) Large vs small genomes in Passiflora: the influence of the mobilome and the satellitome. Planta 253:1–18
Samoluk SS, Chalup LMI, Robledo G, Seijo JG (2015a) Genome sizes in diploid and allopolyploid Arachis L. species (section Arachis). Genet Res Crop Evol 62:747–763
Samoluk SS, Robledo G, Podio M, Chalup L, Ortiz JPA, Pessino SC, Seijo JG (2015b) First insight into divergence, representation and chromosome distribution of reverse transcriptase fragments from L1 retrotransposons in peanut and wild relative species. Genetica 143:113–125
Samoluk SS, Robledo G, Bertioli D, Seijo JG (2017) Evolutionary dynamics of an at-rich satellite DNA and its contribution to karyotype differentiation in wild diploid Arachis species. Mol Genet Genomics 292:283–296
Samoluk SS, Chalup LMI, Chavarro C, Robledo G, Bertioli DJ, Jackson SA, Seijo G (2019) Heterochromatin evolution in Arachis investigated through genome-wide analysis of repetitive DNA. Planta 249:1405–1415
San Miguel P, Bennetzen JL (1998) Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Ann Bot 82:37–44
Santana SH, Valls JF (2015) Arachis veigae (Fabaceae), the most dispersed wild species of the genus, and yet taxonomically overlooked. Bonplandia 24:139–150
Seijo G, Lavia GI, Fernández A, Krapovickas A, Ducasse D, Moscone EA (2004) Physical mapping of 5S and 18S–25S rRNA genes evidences that Arachis duranensis and A. ipaensis are the wild diploid species involved in the origin of A. hypogaea (Leguminosae). Am J Bot 91:2293–2303
Seijo G, Lavia GI, Fernández A, Krapovickas A, Ducasse D, Bertioli DJ, Moscone EA (2007) Genomic relationships between the cultivated peanut (Arachis hypogaea-Leguminosae) and its close relatives revealed by double GISH. Am J Bot 94:1963–1971
Seijo JG, Kovalsky IE, Chalup LMI, Samoluk SS, Fávero A, Robledo G (2018) Karyotype stability and genome specific nucleolar dominance in peanut, its wild 4× ancestor and in a synthetic AABB polyploidy. Crop Sci 58:1671–1683
Seijo GJ, Atahuachi M, Simpson CE, Krapovickas A (2021) Arachis inflata. Bonplandia 30:169–174
Shirasawa K, Koilkonda P, Aoki K, Hirakawa H, Tabata S, Watanabe M, Hasegawa M, Kiyoshima H, Suzuki S, Kuwata C, Naito Y, Kuboyama T, Nakaya A, Sasamoto S, Watanabe A, Kato M, Kawashima K, Kishida Y, Kohara M, Kurabayashi A, Takahashi C, Tsuruoka H, Wada T, Isobe S (2012) In silico polymorphism analysis for the development of simple sequence repeat and transposon markers and construction of linkage map in cultivated peanut. BMC Plant Biol 12:80
Silva Oliveira MA, Nunes T, Dos Santos MA, Ferreira Gomes D, Costa I, Van-Lume B, Marques da Silva SS, Oliveira RS, Simon M, Lima GS, Gissi DS, Almeida CC, Souza G, Marques A (2021) High-throughput genomic data reveal complex phylogenetic relationships in Stylosanthes Sw (Leguminosae). Front Genet 12:727314
Silvestri MC, Ortiz AM, Lavia GI (2014) rDNA loci and heterochromatin positions support a distinct genome type for ‘x = 9 species’ of section Arachis (Arachis, Leguminosae). Plant Syst Evol 301:555–562
Smartt J, Gregory WC, Gregory MP (1978) The genomes of Arachis hypogaea. 1. Cytogenetic studies of putative genome donors. Euphytica 27:665–675
Stalker HT (1991) A new species-section Arachis of peanuts with D genome. Am J Bot 78:630–637
Stalker HT, Dhesi JS, Parry DC, Hahn JH (1991) Cytological and interfertility relationships of Arachis section Arachis. Am J Bot 78:238–246
Staton SE, Burke JM (2015) Evolutionary transitions in the Asteraceae coincide with marked shifts in transposable element abundance. BMC Genomics 16:623
Ugarkovic D, Plohl M (2002) Variation in satellite DNA profiles-causes and effects. EMBO J 2:5955–5959
Usai G, Mascagni F, Natali L, Giordani T, Cavallini A (2017) Comparative genome-wide analysis of repetitive DNA in the genus Populus L. Tree Genet Genomes 13:96
Vaio M, Mazzella C, Porro V, Speranza P, López-Carro B, Estramil E, Folle GA (2007) Nuclear DNA content in allopolyploid species and synthetic hybrids in the grass genus Paspalum. Plant Syst Evol 265:109–121
Valls JFM, Simpson CE (2005) New species of Arachis from Brazil, Paraguay, and Bolivia. Bonplandia 14:35–64
Valls JFM, Simpson CE (2017) A new species of Arachis (Fabaceae) from Mato Grosso, Brazil, related to Arachis matiensis. Bonplandia 26:143–149
Valls JFM, Da Costa LC, Custodio AR (2013) A novel trifoliolate species of Arachis (Fabaceae) and further comments on the taxonomic section Trierectoides. Bonplandia 22:91–97
Vitales D, Garcia S, Dodsworth S (2020) Reconstructing phylogenetic relationships based on repeat sequence similarities. Mol Phylogenet Evol 147:106766
Wang J, Li Y, Li C, Yan C, Zhao X, Yuan C, Sun Q, Shi C, Shan S (2019) Twelve complete chloroplast genomes of wild peanuts: great genetic resources and a better understanding of Arachis phylogeny. BMC Plant Biol 19:1–18
Weiss-Schneeweiss H, Leitch AR, McCann J, Jang TS, Macas J (2015) Employing next generation sequencing to explore the repeat landscape of the plant genome. Next generation sequencing in plant systematics. Regnum Veget 157:155–179
Woo TH, Hong TH, Kim SS, Chung WH, Kang HJ, Kim CB, Seo JM (2007) Repeatome: a database for repeat element comparative analysis in human and chimpanzee. Genomics Inform 5:179–187
Yin D, Wang Y, Zhang X, Ma X, He X, Zhang J (2017) Development of chloroplast genome resources for peanut (Arachis hypogaea L.) and other species of Arachis. Sci Rep 7:1–11
Zhang QJ, Gao LZ (2017) Rapid and recent evolution of LTR retrotransposons drives rice genome evolution during the speciation of AA-genome Oryza species. G3: Genes Genom Genet 7:1875–1885
Zhang L, Xu C, Yu W (2012) Cloning and characterization of chromosomal markers from a Cot-1 library of peanut (Arachis hypogaea L.). Cytogenet Genome Res 137:31–41
Zhang L, Yang X, Tian L, Chen L, Yu W (2016) Identification of peanut (Arachis hypogaea) chromosomes using a fluorescence in situ hybridization system reveals multiple hybridization events during tetraploid peanut formation. New Phytol 211:1424–1439
Acknowledgements
This work was supported by Grants (PICT 2017-3220; PICT 2018-3664) of Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT) and Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Argentina. We thank United States Department of Agriculture (USDA) for providing seeds of some accessions and Center for Applied Genetic Technologies, University of Georgia (Athens, GA, USA), for supporting the genome sequencing.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Anastasios Melis.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Samoluk, S.S., Vaio, M., Ortíz, A.M. et al. Comparative repeatome analysis reveals new evidence on genome evolution in wild diploid Arachis (Fabaceae) species. Planta 256, 50 (2022). https://doi.org/10.1007/s00425-022-03961-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-022-03961-9