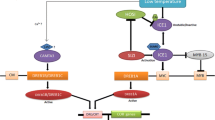
Abstract
Plants can sustain under adverse conditions despite being sessile because of the presence of intricate and sophisticated signaling systems. All the perceived adverse stimuli can activate the signaling pathways, transducing the signals downstream, and bringing about an adaptive response. At the forefront of stress-mediated signaling pathways, transcription factors (TFs) are targeted by the signaling pathways to regulate the gene expression of the respective gene or set of genes. The AP2/ERF superfamily of TFs is important as they can regulate a plethora of plant physiological and developmental responses. This superfamily has four families named AP2, ERF, RAV, and Soloist. The entire superfamily modulates diverse plant responses ranging from development to stress physiology. We briefly discuss our current understanding of this family’s function in modulating a large number of plant physiological responses. One of the important members of this family is ABA REPRESSOR 1 (ABR1, aka ERF111). In this review, we trace and explore the history of ERF111/ABR1 and discuss the protein function—as a transcriptional activator and repressor. Different proteins can modulate ERF111/ABR1, either by binding to its promoter or by affecting it post translationally. ERF111/ABR1 can also modulate plant responses by regulating gene expression. ERF111/ABR1 can connect multiple signaling pathways as it seems to be at the nodal point where several hormonal pathways converge. We have also attempted to explore its possible role as a crosstalk hub and discuss the possible pathways it might regulate as a transcriptional on/off switch.



Similar content being viewed by others
Consent for Publication
All authors give their consent to publish this article.
References
Arce DP, Godoy AV, Tsuda K, Yamazaki K, Valle EM, Iglesias MJ, Di Mauro MF, Casalongue CA (2010) The analysis of an Arabidopsis triple knock-down mutant reveals functions for MBF1 genes under oxidative stress conditions. J Plant Physiol 167:194–200
Bai L, Zhang G, Zhou Y, Zhang Z, Wang W, Du Y, Wu Z, Song CP (2009a) Plasma membrane-associated proline-rich extensin-like receptor kinase 4, a novel regulator of Ca signalling, is required for abscisic acid responses in Arabidopsis thaliana. Plant J 60:314–327
Bai L, Zhou Y, Song CP (2009b) Arabidopsis proline-rich extensin-like receptor kinase 4 modulates the early event toward abscisic acid response in root tip growth. Plant Signal Behav 4:1075–1077
Baillo EH, Kimotho RN, Zhang Z, Xu P (2019) Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes 10:771
Bar-On YM, Phillips R, Milo R (2018) The biomass distribution on Earth. Proc Natl Acad Sci USA 115:6506–6511
Baumler J, Riber W, Klecker M, Muller L, Dissmeyer N, Weig AR, Mustroph A (2019a) AtERF#111/ABR1 is a transcriptional activator involved in the wounding response. Plant J 100:969–990
Bäumler J, Riber W, Klecker M, Müller L, Dissmeyer N, Weig AR, Mustroph A (2019b) At ERF#111/ABR1 is a transcriptional activator involved in the wounding response. Plant J 100:969–990
Bossi F, Cordoba E, Dupre P, Mendoza MS, Roman CS, Leon P (2009) The Arabidopsis ABA-INSENSITIVE (ABI) 4 factor acts as a central transcription activator of the expression of its own gene, and for the induction of ABI5 and SBE2.2 genes during sugar signaling. Plant J 59:359–374
Chandler JW (2018) Class VIIIb APETALA2 ethylene response factors in plant development. Trends Plant Sci 23:151–162
Chen WH, Li PF, Chen MK, Lee YI, Yang CH (2015) FOREVER YOUNG FLOWER negatively regulates ethylene response DNA-binding factors by activating an ethylene-responsive factor to control Arabidopsis floral organ senescence and abscission. Plant Physiol 168:1666–1683
Chen CY, Lin PH, Chen KH, Cheng YS (2020) Structural insights into Arabidopsis ethylene response factor 96 with an extended N-terminal binding to GCC box. Plant Mol Biol 104:483–498
Choi SW, Lee SB, Na YJ, Jeung SG, Kim SY (2017) Arabidopsis MAP3K16 and other salt-inducible MAP3Ks regulate ABA response redundantly. Mol Cells 40:230–242
Ding Y, Liu N, Virlouvet L, Riethoven JJ, Fromm ME, Avramova Z (2013) Four distinct types of dehydration stress memory genes in Arabidopsis thaliana. BMC Plant Biol 13:229
Dobon A, Canet JV, Garcia-Andrade J, Angulo C, Neumetzler L, Persson S, Vera P (2015) Novel disease susceptibility factors for fungal necrotrophic pathogens in Arabidopsis. PLoS Pathog 11:e1004800
Doody E, Zha Y, He J, Poethig RS (2022) The genetic basis of natural variation in the timing of vegetative phase change in Arabidopsis thaliana. Development 149(10):dev200321
Du Y, Scheres B (2017) PLETHORA transcription factors orchestrate de novo organ patterning during Arabidopsis lateral root outgrowth. Proc Natl Acad Sci USA 114:11709–11714
Feng K, Hou XL, Xing GM, Liu JX, Duan AQ, Xu ZS, Li MY, Zhuang J, Xiong AS (2020) Advances in AP2/ERF super-family transcription factors in plant. Crit Rev Biotechnol 40:750–776
Gautam JK, Nandi AK (2018) APD1, the unique member of Arabidopsis AP2 family influences systemic acquired resistance and ethylene-jasmonic acid signaling. Plant Physiol Biochem 133:92–99
Gibbs DJ, Lee SC, Isa NM, Gramuglia S, Fukao T, Bassel GW, Correia CS, Corbineau F, Theodoulou FL, Bailey-Serres J, Holdsworth MJ (2011) Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479:415–418
Giri MK, Swain S, Gautam JK, Singh S, Singh N, Bhattacharjee L, Nandi AK (2014) The Arabidopsis thaliana At4g13040 gene, a unique member of the AP2/EREBP family, is a positive regulator for salicylic acid accumulation and basal defense against bacterial pathogens. J Plant Physiol 171:860–867
Giuntoli B, Perata P (2018) Group VII ethylene response factors in Arabidopsis: regulation and physiological roles. Plant Physiol 176:1143–1155
Gras DE, Vidal EA, Undurraga SF, Riveras E, Moreno S, Dominguez-Figueroa J, Alabadi D, Blázquez MA, Medina J, Gutiérrez RA (2018) SMZ/SNZ and gibberellin signaling are required for nitrate-elicited delay of flowering time in Arabidopsis thaliana. J Exp Bot 69:619–631
Heyman J, Canher B, Bisht A, Christiaens F, De Veylder L (2018) Emerging role of the plant ERF transcription factors in coordinating wound defense responses and repair. J Cell Sci 131(2):jcs208215
Huang PY, Catinot J, Zimmerli L (2016) Ethylene response factors in Arabidopsis immunity. J Exp Bot 67:1231–1241
Huibers RP, de Jong M, Dekter RW, Van den Ackerveken G (2009) Disease-specific expression of host genes during downy mildew infection of Arabidopsis. Mol Plant Microbe Interact 22:1104–1115
Jeena GS, Phukan UP, Singh N, Joshi A, Pandey A, Sharma Y, Tripathi V, Shukla RK (2021) AtERF60 negatively regulates ABR1 to modulate salt, drought, and basal resistance in Arabidopsis. bioRxiv
Jiao Y, Yang H, Ma L, Sun N, Yu H, Liu T, Gao Y, Gu H, Chen Z, Wada M, Gerstein M, Zhao H, Qu LJ, Deng XW (2003) A genome-wide analysis of blue-light regulation of Arabidopsis transcription factor gene expression during seedling development. Plant Physiol 133:1480–1493
Jofuku KD, den Boer BG, Van Montagu M, Okamuro JK (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6:1211–1225
Jones JD, Dangl JL (2006) The plant immune system. Nature 444:323–329
Kagaya Y, Ohmiya K, Hattori T (1999) RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res 27:470–478
Kanwar P, Sanyal SK, Mahiwal S, Ravi B, Kaur K, Fernandes JL, Yadav AK, Tokas I, Srivastava AK, Suprasanna P, Pandey GK (2022) CIPK9 targets VDAC3 and modulates oxidative stress responses in Arabidopsis. Plant J 109:241–260
Kidokoro S, Kim JS, Ishikawa T, Suzuki T, Shinozaki K, Yamaguchi-Shinozaki K (2020) DREB1A/CBF3 is repressed by transgene-induced DNA methylation in the Arabidopsis ice1-1 mutant. Plant Cell 32:1035–1048
Kim KN, Cheong YH, Grant JJ, Pandey GK, Luan S (2003) CIPK3, a calcium sensor-associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. Plant Cell 15:411–423
Krizek BA (2011) Aintegumenta and Aintegumenta-Like6 regulate auxin-mediated flower development in Arabidopsis. BMC Res Notes 4:176
Krizek BA (2022) My favorite flowering image: ‘giant’ Arabidopsis flowers. J Exp Bot 73:3836–3839
Krizek BA, Bantle AT, Heflin JM, Han H, Freese NH, Loraine AE (2021) AINTEGUMENTA and AINTEGUMENTA-LIKE6 directly regulate floral homeotic, growth, and vascular development genes in young Arabidopsis flowers. J Exp Bot 72:5478–5493
Lee SJ, Kang JY, Park HJ, Kim MD, Bae MS, Choi HI, Kim SY (2010) DREB2C interacts with ABF2, a bZIP protein regulating abscisic acid-responsive gene expression, and its overexpression affects abscisic acid sensitivity. Plant Physiol 153:716–727
Lee ZH, Hirakawa T, Yamaguchi N, Ito T (2019) The roles of plant hormones and their interactions with regulatory genes in determining meristem activity. Int J Mol Sci 20:4065
Lewis JD, Guttman DS, Desveaux D (2009) The targeting of plant cellular systems by injected type III effector proteins. Semin Cell Dev Biol 20:1055–1063
Lewis JD, Lee A, Ma W, Zhou H, Guttman DS, Desveaux D (2011) The YopJ superfamily in plant-associated bacteria. Mol Plant Pathol 12:928–937
Li Y, Shu Y, Peng C, Zhu L, Guo G, Li N (2012) Absolute quantitation of isoforms of post-translationally modified proteins in transgenic organism. Mol Cell Proteomics 11:272–285
Li T, Wu XY, Li H, Song JH, Liu JY (2016) A dual-function transcription factor, AtYY1, is a novel negative regulator of the Arabidopsis ABA response network. Mol Plant 9:650–661
Licausi F, Ohme-Takagi M, Perata P (2013) APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. N Phytol 199:639–649
Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10:1391–1406
Liu J, Li J, Wang H, Fu Z, Liu J, Yu Y (2011) Identification and expression analysis of ERF transcription factor genes in Petunia during flower senescence and in response to hormone treatments. J Exp Bot 62:825–840
Liu Y, Ji X, Zheng L, Nie X, Wang Y (2013) Microarray analysis of transcriptional responses to abscisic acid and salt stress in Arabidopsis thaliana. Int J Mol Sci 14:9979–9998
Liu WC, Song RF, Qiu YM, Zheng SQ, Li TT, Wu Y, Song CP, Lu YT, Yuan HM (2022) Sulfenylation of ENOLASE2 facilitates H2O2-conferred freezing tolerance in Arabidopsis. Dev Cell 57:1883–1898
Luo X, Xu J, Zheng C, Yang Y, Wang L, Zhang R, Ren X, Wei S, Aziz U, Du J, Liu W, Tan W, Shu K (2022) Abscisic acid inhibits primary root growth by impairing ABI4-mediated cell cycle and auxin biosynthesis. Plant Physiol 191:265–279
Maes T, Van de Steene N, Zethof J, Karimi M, D’Hauw M, Mares G, Van Montagu M, Gerats T (2001) Petunia Ap2-like genes and their role in flower and seed development. Plant Cell 13:229–244
Mao JL, Miao ZQ, Wang Z, Yu LH, Cai XT, Xiang CB (2016) Arabidopsis ERF1 mediates cross-talk between ethylene and auxin biosynthesis during primary root elongation by regulating ASA1 expression. PLoS Genet 12:e1005760
Mena M, Cejudo FJ, Isabel-Lamoneda I, Carbonero P (2002) A role for the DOF transcription factor BPBF in the regulation of gibberellin-responsive genes in barley aleurone. Plant Physiol 130:111–119
Miret JA, Munne-Bosch S, Dijkwel PP (2018) ABA signalling manipulation suppresses senescence of a leafy vegetable stored at room temperature. Plant Biotechnol J 16:530–544
Mishra M, Kanwar P, Singh A, Pandey A, Kapoor S, Pandey GK (2013) Plant omics: genome-wide analysis of ABA repressor1 (ABR1) related genes in rice during abiotic stress and development. OMICS 17:439–450
Mizoi J, Kanazawa N, Kidokoro S, Takahashi F, Qin F, Morimoto K, Shinozaki K, Yamaguchi-Shinozaki K (2019) Heat-induced inhibition of phosphorylation of the stress-protective transcription factor DREB2A promotes thermotolerance of Arabidopsis thaliana. J Biol Chem 294:902–917
Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140:411–432
Nurnberger T, Brunner F (2002) Innate immunity in plants and animals: emerging parallels between the recognition of general elicitors and pathogen-associated molecular patterns. Curr Opin Plant Biol 5:318–324
Ohme-Takagi M, Shinshi H (1995) Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7:173–182
Okamuro JK, Caster B, Villarroel R, Van Montagu M, Jofuku KD (1997) The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc Natl Acad Sci USA 94:7076–7081
Osnato M, Cereijo U, Sala J, Matías-Hernández L, Aguilar-Jaramillo AE, Rodríguez-Goberna MR, Riechmann JL, Rodríguez-Concepción M, Pelaz S (2021) The floral repressors TEMPRANILLO1 and 2 modulate salt tolerance by regulating hormonal components and photo-protection in Arabidopsis. Plant J 105:7–21
Pandey GK, Cheong YH, Kim KN, Grant JJ, Li L, Hung W, D’Angelo C, Weinl S, Kudla J, Luan S (2004) The calcium sensor calcineurin B-like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. Plant Cell 16:1912–1924
Pandey GK, Grant JJ, Cheong YH, Kim BG, Li L, Luan S (2005) ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiol 139:1185–1193
Pandey GK, Grant JJ, Cheong YH, Kim BG, le Li G, Luan S (2008) Calcineurin-B-like protein CBL9 interacts with target kinase CIPK3 in the regulation of ABA response in seed germination. Mol Plant 1:238–248
Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, Pilgrim M, Broun P, Zhang JZ, Ghandehari D, Sherman BK, Yu G (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290:2105–2110
Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290:998–1009
Sanyal SK, Kanwar P, Samtani H, Kaur K, Jha SK, Pandey GK (2017a) Alternative splicing of CIPK3 results in distinct target selection to propagate ABA signaling in Arabidopsis. Front Plant Sci 8:1924
Sanyal SK, Kanwar P, Yadav AK, Sharma C, Kumar A, Pandey GK (2017b) Arabidopsis CBL interacting protein kinase 3 interacts with ABR1, an APETALA2 domain transcription factor, to regulate ABA responses. Plant Sci 254:48–59
Sanyal SK, Mahiwal S, Nambiar DM, Pandey GK (2020) CBL-CIPK module-mediated phosphoregulation: facts and hypothesis. Biochem J 477:853–871
Sato H, Takasaki H, Takahashi F, Suzuki T, Iuchi S, Mitsuda N, Ohme-Takagi M, Ikeda M, Seo M, Yamaguchi-Shinozaki K, Shinozaki K (2018) Arabidopsis thaliana NGATHA1 transcription factor induces ABA biosynthesis by activating NCED3 gene during dehydration stress. Proc Natl Acad Sci USA 115:E11178–E11187
Schreiber KJ, Hassan JA, Lewis JD (2021) Arabidopsis Abscisic Acid Repressor 1 is a susceptibility hub that interacts with multiple Pseudomonas syringae effectors. Plant J 105:1274–1292
Serra TS, Figueiredo DD, Cordeiro AM, Almeida DM, Lourenco T, Abreu IA, Sebastian A, Fernandes L, Contreras-Moreira B, Oliveira MM, Saibo NJ (2013) OsRMC, a negative regulator of salt stress response in rice, is regulated by two AP2/ERF transcription factors. Plant Mol Biol 82:439–455
Sharma P, Watts A, Kumar V, Srinivasan R, Siwach P (2018) Cloning, characterization and expression analysis of APETALA2 genes of Brassica juncea (L.) Czern. Indian J Exp Biol 56:604–610
Shikata M, Matsuda Y, Ando K, Nishii A, Takemura M, Yokota A, Kohchi T (2004) Characterization of Arabidopsis ZIM, a member of a novel plant-specific GATA factor gene family. J Exp Bot 55:631–639
Shinozaki K, Yamaguchi-Shinozaki K (2000) Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol 3:217–223
Shu K, Zhou W, Yang W (2018) APETALA 2-domain-containing transcription factors: focusing on abscisic acid and gibberellins antagonism. N Phytol 217:977–983
Singh K, Chandra A (2021) DREBs-potential transcription factors involve in combating abiotic stress tolerance in plants. Biologia 76:3043–3055
Song L, Huang SC, Wise A, Castanon R, Nery JR, Chen H, Watanabe M, Thomas J, Bar-Joseph Z, Ecker JR (2016) A transcription factor hierarchy defines an environmental stress response network. Science 354:aag1550
Song Y, Zhang X, Li M, Yang H, Fu D, Lv J, Ding Y, Gong Z, Shi Y, Yang S (2021) The direct targets of CBFs: in cold stress response and beyond. J Integr Plant Biol 63:1874–1887
Sun L, Dong X, Song X (2023) PtrABR1 increases tolerance to drought stress by enhancing lateral root formation in Populus trichocarpa. Int J Mol Sci 24:13748
To A, Joubès J, Barthole G, Lécureuil A, Scagnelli A, Jasinski S, Lepiniec L, Baud S (2012) WRINKLED transcription factors orchestrate tissue-specific regulation of fatty acid biosynthesis in Arabidopsis. Plant Cell 24:5007–5023
van Veen H, Vashisht D, Akman M, Girke T, Mustroph A, Reinen E, Hartman S, Kooiker M, van Tienderen P, Schranz ME, Bailey-Serres J, Voesenek LA, Sasidharan R (2016) Transcriptomes of eight Arabidopsis thaliana accessions reveal core conserved, genotype- and organ-specific responses to flooding stress. Plant Physiol 172:668–689
Vonapartis E, Mohamed D, Li J, Pan W, Wu J, Gazzarrini S (2022) CBF4/DREB1D represses XERICO to attenuate ABA, osmotic and drought stress responses in Arabidopsis. Plant J 110:961–977
Wang Y, Shirakawa M, Ito T (2022) Arrest, senescence, and death of shoot apical stem cells in Arabidopsis thaliana. Plant Cell Physiol 64:284–290
Wind JJ, Peviani A, Snel B, Hanson J, Smeekens SC (2013) ABI4: versatile activator and repressor. Trends Plant Sci 18:125–132
Xiao S, Hu Q, Zhang X, Si H, Liu S, Chen L, Chen K, Berne S, Yuan D, Lindsey K, Zhang X, Zhu L (2021) Orchestration of plant development and defense by indirect crosstalk of salicylic acid and brassinosteroid signaling via transcription factor GhTINY2. J Exp Bot 72:4721–4743
Xie Z, Nolan TM, Jiang H, Yin Y (2019) AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front Plant Sci 10:228
Xin XF, Kvitko B, He SY (2018) Pseudomonas syringae: what it takes to be a pathogen. Nat Rev Microbiol 16:316–328
Xu Y, Qian Z, Zhou B, Wu G (2019) Age-dependent heteroblastic development of leaf hairs in Arabidopsis. N Phytol 224:741–748
Ye BB, Shang GD, Pan Y, Xu ZG, Zhou CM, Mao YB, Bao N, Sun L, Xu T, Wang JW (2020) AP2/ERF transcription factors integrate age and wound signals for root regeneration. Plant Cell 32:226–241
Yeung E, van Veen H, Vashisht D, Sobral Paiva AL, Hummel M, Rankenberg T, Steffens B, Steffen-Heins A, Sauter M, de Vries M, Schuurink RC, Bazin J, Bailey-Serres J, Voesenek L, Sasidharan R (2018) A stress recovery signaling network for enhanced flooding tolerance in Arabidopsis thaliana. Proc Natl Acad Sci USA 115:E6085–E6094
Yuan L (2020) Clustered ERF transcription factors: not all created equal. Plant Cell Physiol 61:1025–1027
Zhang Y, Zhu J, Khan M, Wang Y, Xiao W, Fang T, Qu J, Xiao P, Li C, Liu JH (2023) Transcription factors ABF4 and ABR1 synergistically regulate amylase-mediated starch catabolism in drought tolerance. Plant Physiol 191:591–609
Zhao J, Shi M, Yu J, Guo C (2022) SPL9 mediates freezing tolerance by directly regulating the expression of CBF2 in Arabidopsis thaliana. BMC Plant Biol 22:59
Zhu JK (2016) Abiotic stress signaling and responses in plants. Cell 167:313–324
Zhu Y, Wang K, Wu C, Zhao Y, Yin X, Zhang B, Grierson D, Chen K, Xu C (2019) Effect of ethylene on cell wall and lipid metabolism during alleviation of postharvest chilling injury in peach. Cells 8:1612
Zipfel C (2009) Early molecular events in PAMP-triggered immunity. Curr Opin Plant Biol 12:414–420
Zipfel C, Robatzek S (2010) Pathogen-associated molecular pattern-triggered immunity: Veni, Vidi…? Plant Physiol 154:551–554
Zumajo-Cardona C, Pabón-Mora N, Ambrose BA (2021) The evolution of euAPETALA2 genes in vascular plants: from plesiomorphic roles in sporangia to acquired functions in ovules and fruits. Mol Biol Evol 38:2319–2336
Acknowledgements
We are thankful to the Department of Biotechnology (DBT), Government of India; Science and Engineering Research Board (SERB), Government of India; Council for Scientific and Industrial Research (CSIR), and Government of India; Delhi University (IoE/FRP grant), India for research funding in our lab.
Author information
Authors and Affiliations
Contributions
Girdhar K. Pandey contributed to Conceptualization, Supervision, Funding acquisition, and Writing, reviewing, and editing of the manuscript. Sibaji K. Sanyal contributed to Investigation, Methodology, Writing of the original draft, and Writing, reviewing, and editing of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this article.
Additional information
Handling Editor: Ashish Srivastava.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sanyal, S.K., Pandey, G.K. ERF111/ABR1: An AP2 Domain Transcription Factor Serving as a Hub for Multiple Signaling Pathways. J Plant Growth Regul (2024). https://doi.org/10.1007/s00344-023-11225-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00344-023-11225-3