Abstract
Key message
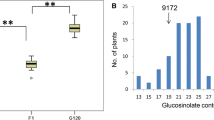
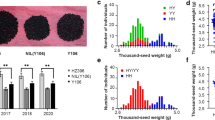
By integrating QTL fine mapping and transcriptomics, a candidate gene responsible for oil content in rapeseed was identified. The gene is anticipated to primarily function in photosynthesis and photosystem metabolism pathways.
Abstract
Brassica napus is one of the most important oil crops in the world, and enhancing seed oil content is an important goal in its genetic improvement. However, the underlying genetic basis for the important trait remains poorly understood in this crop. We previously identified a major locus, OILA5 responsible for seed oil content on chromosome A5 through genome-wide association study. To better understand the genetics of the QTL, we performed fine mapping of OILA5 with a double haploid population and a BC3F2 segregation population consisting of 6227 individuals. We narrowed down the QTL to an approximate 43 kb region with twelve annotated genes, flanked by markers ZDM389 and ZDM337. To unveil the potential candidate gene responsible for OILA5, we integrated fine mapping data with transcriptome profiling using high and low oil content near-isogenic lines. Among the candidate genes, BnaA05G0439400ZS was identified with high expression levels in both seed and silique tissues. This gene exhibited homology with AT3G09840 in Arabidopsis that was annotated as cell division cycle 48. We designed a site-specific marker based on resequencing data and confirmed its effectiveness in both natural and segregating populations. Our comprehensive results provide valuable genetic information not only enhancing our understanding of the genetic control of seed oil content but also novel germplasm for advancing high seed oil content breeding in B. napus and other oil crops.







Similar content being viewed by others
Data availability
All the raw sequencing data generated during this study are available in the Genome Sequence Archive (https://bigd.big.ac.cn/gsa/) with Bio-project ID PRJCA017272. Data will be made publicly accessible after publication of the manuscript.
References
Behnke N, Suprianto E, Mollers C (2018) A major QTL on chromosome C05 significantly reduces acid detergent lignin (ADL) content and increases seed oil and protein content in oilseed rape (Brassica napus L.). Theor Appl Genet 131(11):2477–2492
Bodnar N, Rapoport T (2017) Toward an understanding of the Cdc48/p97 ATPase. F1000Res 6:1318
Chalhoub B, Denoeud F, Liu S, Parkin IA, Tang H, Wang X, Chiquet J, Belcram H, Tong C, Samans B, Correa M, Da Silva C, Just J, Falentin C, Koh CS, Le Clainche I, Bernard M, Bento P, Noel B, Labadie K, Alberti A, Charles M, Arnaud D, Guo H, Daviaud C, Alamery S, Jabbari K, Zhao M, Edger PP, Chelaifa H, Tack D, Lassalle G, Mestiri I, Schnel N, Le Paslier MC, Fan G, Renault V, Bayer PE, Golicz AA, Manoli S, Lee TH, Thi VH, Chalabi S, Hu Q, Fan C, Tollenaere R, Lu Y, Battail C, Shen J, Sidebottom CH, Wang X, Canaguier A, Chauveau A, Berard A, Deniot G, Guan M, Liu Z, Sun F, Lim YP, Lyons E, Town CD, Bancroft I, Wang X, Meng J, Ma J, Pires JC, King GJ, Brunel D, Delourme R, Renard M, Aury JM, Adams KL, Batley J, Snowdon RJ, Tost J, Edwards D, Zhou Y, Hua W, Sharpe AG, Paterson AH, Guan C, Wincker P (2014) Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345(6199):950–953
Chao H, Guo L, Zhao W, Li H, Li M (2022) A major yellow-seed QTL on chromosome A09 significantly increases the oil content and reduces the fiber content of seed in Brassica napus. Theor Appl Genet 135(4):1293–1305
Chen Y, Qi L, Zhang X, Huang J, Wang J, Chen H, Ni X, Xu F, Dong Y, Xu H, Zhao J (2013) Characterization of the quantitative trait locus OilA1 for oil content in Brassica napus. Theor Appl Genet 126(10):2499–2509
Chen J, Wang B, Zhang Y, Yue X, Li Z, Liu K (2017) High-density ddRAD linkage and yield-related QTL mapping delimits a chromosomal region responsible for oil content in rapeseed (Brassica napus L.). Breed Sci 67(3):296–306
Cui Y, Zeng X, Xiong Q, Wei D, Liao J, Xu Y, Chen G, Zhou Y, Dong H, Wan H, Liu Z, Li J, Guo L, Jung C, He Y, Qian W (2021) Combining quantitative trait locus and co-expression analysis allowed identification of new candidates for oil accumulation in rapeseed. J Exp Bot 72(5):1649–1660
Ding LN, Guo XJ, Li M, Fu ZL, Yan SZ, Zhu KM, Wang Z, Tan XL (2019) Improving seed germination and oil contents by regulating the GDSL transcriptional level in Brassica napus. Plant Cell Rep 38(2):243–253
Guo Y, Si P, Wang N, Wen J, Yi B, Ma C, Tu J, Zou J, Fu T, Shen J (2017) Genetic effects and genotype x environment interactions govern seed oil content in Brassica napus L. BMC Genet 18(1):1
Hajduch M, Casteel JE, Hurrelmeyer KE, Song Z, Agrawal GK, Thelen JJ (2006) Proteomic analysis of seed filling in Brassica napus. Developmental characterization of metabolic isozymes using high-resolution two-dimensional gel electrophoresis. Plant Physiol 141(1):32–46
Huang KL, Zhang ML, Ma GJ, Wu H, Wu XM, Ren F, Li XB (2017) Transcriptome profiling analysis reveals the role of silique in controlling seed oil content in Brassica napus. PLoS ONE 12(6):e0179027
Jia Y, Yao M, He X, Xiong X, Guan M, Liu Z, Guan C, Qian L (2022) Transcriptome and regional association analyses reveal the effects of oleosin genes on the accumulation of oil content in Brassica napus. Plants 11(22):3140
Jiang C, Shi J, Li R, Long Y, Wang H, Li D, Zhao J, Meng J (2014) Quantitative trait loci that control the oil content variation of rapeseed (Brassica napus L.). Theor Appl Genet 127(4):957–968
Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK (2012) VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22(3):568–576
Langmead B, Salzberg S (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9(4):357–359
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newberg LA (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1(2):174–181
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Li J, Yuan J, Li Y, Sun H, Ma T, Huai J, Yang W, Zhang W, Lin R (2022) The CDC48 complex mediates ubiquitin-dependent degradation of intra-chloroplast proteins in plants. Cell Rep 39(2):110664
Li Y, Tan Z, Zeng C, Xiao M, Lin S, Yao W, Li Q, Guo L, Lu S (2023) Regulation of seed oil accumulation by lncRNAs in Brassica napus. Biotechnol Biofuels Bioprod 16(1):22
Ling Q, Broad W, Trosch R, Topel M, Demiral Sert T, Lymperopoulos P, Baldwin A, Jarvis RP (2019) Ubiquitin-dependent chloroplast-associated protein degradation in plants. Science 363(6429):eaav4467
Liu S, Fan C, Li J, Cai G, Yang Q, Wu J, Yi X, Zhang C, Zhou Y (2016) A genome-wide association study reveals novel elite allelic variations in seed oil content of Brassica napus. Theor Appl Genet 129(6):1203–1215
Liu J, Hao W, Liu J, Fan S, Zhao W, Deng L, Wang X, Hu Z, Hua W, Wang H (2019) A novel chimeric mitochondrial gene confers cytoplasmic effects on seed oil content in polyploid rapeseed (Brassica napus L.). Mol Plant 12(4):582–596
Love MI, Huber W, Anders S (2019) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15(12):550
Lu C, Napier JA, Clemente TE, Cahoon EB (2011) New frontiers in oilseed biotechnology: meeting the global demand for vegetable oils for food, feed, biofuel, and industrial applications. Curr Opin Biotechnol 22(2):252–259
Namazkar S, Stockmarr A, Frenck G, Egsgaard H, Terkelsen T, Mikkelsen T, Ingvordsen CH, Jorgensen RB (2016) Concurrent elevation of CO2, O3 and temperature severely affects oil quality and quantity in rapeseed. J Exp Bot 67(14):4117–4125
Patel RK, Jain M (2012) NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS ONE 7:e30619
Qiu D, Morgan C, Shi J, Long Y, Liu J, Li R, Zhuang X, Wang Y, Tan X, Dietrich E, Weihmann T, Everett C, Vanstraelen S, Beckett P, Fraser F, Trick M, Barnes S, Wilmer J, Schmidt R, Li J, Li D, Meng J, Bancroft I (2006) A comparative linkage map of oilseed rape and its use for QTL analysis of seed oil and erucic acid content. Theor Appl Genet 114(1):67–80
Schilbert HM, Holzenkamp K, Viehöver P, Holtgräwe D, Möllers C (2023) Homoeologous non-reciprocal translocation explains a major QTL for seed lignin content in oilseed rape (Brassica napus L.). Theor Appl Genet 136(8):172
Shahid M, Cai G, Zu F, Zhao Q, Qasim MU, Hong Y, Fan C, Zhou Y (2019) Comparative transcriptome analysis of developing seeds and silique wall reveals dynamic transcription networks for effective oil production in Brassica napus L. Int J Mol Sci 20(8):1982
Shi J, Lang C, Wang F, Wu X, Liu R, Zheng T, Zhang D, Chen J, Wu G (2017) Depressed expression of FAE1 and FAD2 genes modifies fatty acid profiles and storage compounds accumulation in Brassica napus seeds. Plant Sci 263:177–182
Song G, Li X, Munir R, Khan AR, Azhar W, Yasin MU, Jiang Q, Bancroft I, Gan Y (2020a) The WRKY6 transcription factor affects seed oil accumulation and alters fatty acid compositions in Arabidopsis thaliana. Physiol Plant 169(4):612–624
Song JM, Guan Z, Hu J, Guo C, Yang Z, Wang S, Liu D, Wang B, Lu S, Zhou R, Xie WZ, Cheng Y, Zhang Y, Liu K, Yang QY, Chen LL, Guo L (2020b) Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat Plants 6(1):34–45
Song JM, Liu DX, Xie WZ, Yang Z, Guo L, Liu K, Yang QY, Chen LL (2021) BnPIR: Brassica napus pan-genome information resource for 1689 accessions. Plant Biotechnol J 19(3):412–414
Stolz A, Hilt W, Buchberger A, Wolf DH (2011) Cdc48: a power machine in protein degradation. Trends Biochem Sci 36(10):515–523
Sun F, Liu J, Hua W, Sun X, Wang X, Wang H (2016) Identification of stable QTLs for seed oil content by combined linkage and association mapping in Brassica napus. Plant Sci 252:388–399
Sun F, Fan G, Hu Q, Zhou Y, Guan M, Tong C, Li J, Du D, Qi C, Jiang L, Liu W, Huang S, Chen W, Yu J, Mei D, Meng J, Zeng P, Shi J, Liu K, Wang X, Wang X, Long Y, Liang X, Hu Z, Huang G, Dong C, Zhang H, Li J, Zhang Y, Li L, Shi C, Wang J, Lee SM, Guan C, Xu X, Liu S, Liu X, Chalhoub B, Hua W, Wang H (2017) The high-quality genome of Brassica napus cultivar ‘ZS11’ reveals the introgression history in semi-winter morphotype. Plant J 92(3):452–468
Tang S, Guo N, Tang Q, Peng F, Liu Y, Xia H, Lu S, Guo L (2022a) Pyruvate transporter BnaBASS2 impacts seed oil accumulation in Brassica napus. Plant Biotechnol J 20(12):2406–2417
Tang S, Peng F, Tang Q, Liu Y, Xia H, Yao X, Lu S, Guo L (2022b) BnaPPT1 is essential for chloroplast development and seed oil accumulation in Brassica napus. J Adv Res 42:29–40
Tang S, Zhao H, Lu S, Yu L, Zhang G, Zhang Y, Yang QY, Zhou Y, Wang X, Ma W, Xie W, Guo L (2021) Genome- and transcriptome-wide association studies provide insights into the genetic basis of natural variation of seed oil content in Brassica napus. Mol Plant 14(3):470–487
Wang X, Wang H, Long Y, Li D, Yin Y, Tian J, Chen L, Liu L, Zhao W, Zhao Y, Yu L, Li M (2013) Identification of QTLs associated with oil content in a high-oil Brassica napus cultivar and construction of a high-density consensus map for QTLs comparison in B. napus. PLoS ONE 8(12):e80569
Wang H, Wang Q, Pak H, Yan T, Chen M, Chen X, Wu D, Jiang L (2021) Genome-wide association study reveals a patatin-like lipase relating to the reduction of seed oil content in Brassica napus. BMC Plant Biol 21(1):6
Wei W, Li G, Jiang X, Wang Y, Ma Z, Niu Z, Wang Z, Geng X (2018) Small RNA and degradome profiling involved in seed development and oil synthesis of Brassica napus. PLoS ONE 13(10):e0204998
Wu J, Chen P, Zhao Q, Cai G, Hu Y, Xiang Y, Yang Q, Wang Y, Zhou Y (2019) Co-location of QTL for Sclerotinia stem rot resistance and flowering time in Brassica napus. Crop J 7:227–237
Xiao Z, Tang F, Zhang L, Li S, Wang S, Huo Q, Yang B, Zhang C, Wang D, Li Q, Wei L, Guo T, Qu C, Lu K, Zhang Y, Guo L, Li J, Li N (2021) The Brassica napus fatty acid exporter FAX1-1 contributes to biological yield, seed oil content, and oil quality. Biotechnol Biofuels 14(1):190
Xiao Z, Zhang C, Tang F, Yang B, Zhang L, Liu J, Huo Q, Wang S, Li S, Wei L, Du H, Qu C, Lu K, Li J, Li N (2019) Identification of candidate genes controlling oil content by combination of genome-wide association and transcriptome analysis in the oilseed crop Brassica napus. Biotechnol Biofuels 12:216
Xiao Z, Zhang C, Qu C, Wei L, Zhang L, Yang B, Lu K, Li J (2022) Identification of candidate genes regulating seed oil content by QTL mapping and transcriptome sequencing in Brassica napus. Front Plant Sci 13:1067121
Xu HM, Kong XD, Chen F, Huang JX, Lou XY, Zhao JY (2015) Transcriptome analysis of Brassica napus pod using RNA-Seq and identification of lipid-related candidate genes. BMC Genomics 16:858
Yao M, Guan M, Yang Q, Huang L, Xiong X, Jan HU, Voss-Fels KP, Werner CR, He X, Qian W, Snowdon RJ, Guan C, Hua W, Qian L (2021) Regional association analysis coupled with transcriptome analyses reveal candidate genes affecting seed oil accumulation in Brassica napus. Theor Appl Genet 134(5):1545–1555
Zafar S, Li YL, Li NN, Zhu KM, Tan XL (2019) Recent advances in enhancement of oil content in oilseed crops. J Biotechnol 301:35–44
Zhang K, He J, Yin Y, Chen K, Deng X, Yu P, Li H, Zhao W, Yan S, Li M (2022) Lysophosphatidic acid acyltransferase 2 and 5 commonly, but differently, promote seed oil accumulation in Brassica napus. Biotechnol Biofuels Bioprod 15(1):83
Zhao J, Huang J, Chen F, Xu F, Ni X, Xu H, Wang Y, Jiang C, Wang H, Xu A, Huang R, Li D, Meng J (2012) Molecular mapping of Arabidopsis thaliana lipid-related orthologous genes in Brassica napus. Theor Appl Genet 124(2):407–421
Zhao Q, Wu J, Cai G, Yang Q, Shahid M, Fan C, Zhang C, Zhou Y (2019) A novel quantitative trait locus on chromosome A9 controlling oleic acid content in Brassica napus. Plant Biotechnol J 17(12):2313–2324
Zhao C, Xie M, Liang L, Yang L, Han H, Qin X, Zhao J, Hou Y, Dai W, Du C, Xiang Y, Liu S, Huang X (2022) Genome-wide association analysis combined with quantitative trait loci mapping and dynamic transcriptome unveil the genetic control of seed oil content in Brassica napus L. Front Plant Sci 13:929197
Zhou Z, Lin B, Tan J, Hao P, Hua S, Deng Z (2022) Tandem mass tag-based quantitative proteomics reveals implication of a late embryogenesis abundant protein (BnLEA57) in seed oil accumulation in Brassica napus L. Front Plant Sci 13:907244
Zhu K, Li N, Zheng X, Sarwar R, Li Y, Cao J, Wang Z, Tan X (2023) Overexpression the BnLACS9 could increase the chlorophyll and oil content in Brassica napus. Biotechnol Biofuels Bioprod 16(1):3
Zou J, Jiang C, Cao Z, Li R, Long Y, Chen S, Meng J (2010) Association mapping of seed oil content in Brassica napus and comparison with quantitative trait loci identified from linkage mapping. Genome 53(11):908–916
Funding
This study was supported by the National Key Research and Development Program (2017YFE0104800).
Author information
Authors and Affiliations
Contributions
YZ conceived the study. QZ and JW designed the experiments. JW, KY, CZ, and CF helped in data analysis. LL, MS, and MQ helped in data collection. QZ, JW, and YZ wrote and revised the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Ethical approval and consent to participate
Not applicable.
Additional information
Communicated by Annaliese S Mason.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, Q., Wu, J., Lan, L. et al. Fine mapping and candidate gene analysis of a major QTL for oil content in the seed of Brassica napus. Theor Appl Genet 136, 256 (2023). https://doi.org/10.1007/s00122-023-04501-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00122-023-04501-z