Abstract
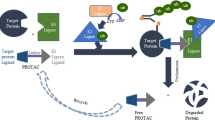
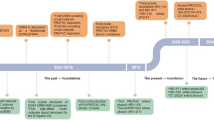
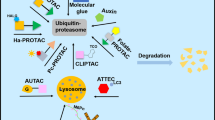
Traditional targets and modalities that have provided rich substrates for medicine design and decades of productive industrial and academic research are not the primary drug discovery engines of the future. There is a steady shift in the target and modality landscape. Novel modalities such as protein degradation are an intense and hot area of research. The explosion of proximity induced ubiquitination of targets and proteasome or lysosome mediated degradation by expropriating E3 ligases (PROTACS, LYTACS) has inspired a number of other proximity induced post-translational modifications such as de-ubiquitination (DUBTACS), phosphorylation (PHICS), de-phosphorylation (PHORCS), acetylation (AceTAGs) to name a few. This review article of emerging TACs (targeting chimeras) provides a sampling of some of the chimeric molecules that preempt these cellular events and highlight opportunities and challenges for prospective medicine design and scientific research.






Similar content being viewed by others
References
Walsh C, Garneau-Tsodikova S, Gatto GJ. Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem Int Ed Engl. 2005;44:7342–72.
Lai AC, Crews CM. Induced protein degradation: an emerging drug discovery paradigm. Nat Rev Drug Discov. 2017;16:101–14.
Ottis P, Crews C. Proteolysis-targeting chimeras: induced protein degradation as a therapeutic strategy. ACS Chem Biol. 2017;12:892–8.
Bekes M, Langley DR, Crews CM. PROTAC targeted protein degraders: the past is prologue. Nat Rev Drug Discov. 2022;21:181–200.
Li X, Pu W, Zheng Q, Ai M, Chen S, Peng Y. Proteolysis-targeting chimeras (PROTACs) in cancer therapy. Mol Cancer. 2022;21:99.
Zhao L, Zhao J, Zhong K, Tong A, Jia D. Targeted protein degradation: mechanisms, strategies and application. Signal Transduct Target Ther. 2022;7:113.
Bondeson DP, Smith BE, Burslem GM, Buhimschi AD, Hines J, Jaime-Figueroa S, et al. Lessons in PROTAC design from selective degradation with a promiscuous warhead. Cell Chem Biol. 2018;25:78–87.e5.
Schapira M, Calabrese MF, Bullock AN, Crews CM. Targeted protein degradation: expanding the toolbox. Nat Rev Drug Discov. 2019;18:949–63.
Kim C, Wang XD, Liu Z, Zha S, Yu Y. Targeting scaffolding functions of enzymes using PROTAC approaches. Biochemistry. 2023;62:561–3.
Imaide S, Riching KM, Makukhin N, Vetma V, Whitworth C, Hughes SJ, et al. Trivalent PROTACs enhance protein degradation via combined avidity and cooperativity. Nat Chem Biol. 2021;17:1157–67.
Kiessling LL, Gestwicki JE, Strong LE. Synthetic multivalent ligands as probes of signal transduction. Angew Chem Int Ed. 2006;45:2348–68.
Zengerle M, Chan K-H, Ciulli A. Selective small molecule induced degradation of the BET bromodomain protein BRD4. ACS Chem Biol. 2015;10:1770–7.
Demars KM, Yang C, Castro-Rivera CI, Candelario-Jalil E. Selective degradation of BET proteins with dBET1, a proteolysis-targeting chimera, potently reduces pro-inflammatory responses in lipopolysaccharide-activated microglia. Biochem. Biophys. Res Commun. 2018;497:410–5.
Lu J, Qian Y, Altieri M, Dong H, Wang J, Raina K, et al. Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chem Biol. 2015;22:755–63.
Douglass EF Jr., Miller CJ, Sparer G, Shapiro H, Spiegel DA. A comprehensive mathematical model for three-body binding equilibria. J Am Chem Soc. 2013;135:6092–9.
Henning NJ, Boike L, Spradlin JN, Ward CC, Liu G, Zhang E, et al. Deubiquitinase-targeting chimeras for targeted protein stabilization. Nat Chem Biol. 2022;18:412–21.
Liu J, Yu X, Chen H, Kaniskan HU, Xie L, Chen X, et al. TF-DUBTACs stabilize tumor suppressor transcription factors. J Am Chem Soc. 2022;144:12934–41.
Yamazoe S, Tom J, Fu Y, Wu W, Zeng L, Sun C, et al. Heterobifunctional molecules induce dephosphorylation of kinases-A proof of concept study. J Med Chem. 2020;63:2807–13.
Hu Z, Chen PH, Li W, Douglas T, Hines J, Liu Y, et al. Targeted dephosphorylation of tau by phosphorylation targeting chimeras (PhosTACs) as a therapeutic modality. J Am Chem Soc. 2023. https://doi.org/10.1021/jacs.2c11706. Online ahead of print.
Zheng J, Tian N, Liu F, Zhang Y, Su J, Gao Y, et al. A novel dephosphorylation targeting chimera selectively promoting tau removal in tauopathies. Signal Transduct Target Ther. 2021;6:269–78.
Chen P-H, Hu Z, An E, Okeke I, Zheng S, Luo X, et al. Modulation of phosphoprotein activity by phosphorylation targeting chimeras (PhosTACs). ACS Chem Biol. 2021;16:2808–15.
Zhang Q, Wu X, Zhang H, Wu Q, Fu M, Hua L, et al. Protein phosphatase 5‐recruiting chimeras for accelerating apoptosis-signal-regulated kinase 1 dephosphorylation with antiproliferative activity. J Am Chem Soc. 2023;145:1118–28.
Testa A, Hughes SJ, Lucas X, Wright JE, Ciulli A. Structure-based DesignofaMacrocyclic PROTAC. Angew Chem Int Ed. 2020;59:1727–34.
Bond MJ, Chu L, Nalawansha DA, Li K, Crews CM. Targeted degradation of oncogenic KRASG12C by VHL-recruiting PROTACs. ACS Cent Sci. 2020;6:1367–75.
Buhimschi AD, Armstrong HA, Toure M, Jaime-Figueroa S, Chen TL, Lehman AM, et al. Targeting the C481S ibrutinib-resistance mutation in bruton’s tyrosine kinase using PROTAC-mediated degradation. Biochemistry. 2018;57:3564–75.
Sachini U, Siriwardena DNPMG, Shoba VM, Lai S, Shi M, Wu P, Chaudhary SK, Schreiber SL, Choudhary A. Phosphorylation-inducing chimeric small molecules. J Am Chem Soc. 2020;142:14052–7.
Pergu R, Shoba VM, Chaudhary SK, Godage DNPM, Deb A, Singha S, et al. Development and applications of chimera platforms for tyrosine phosphorylation. bioRxiv. 2023. https://doi.org/10.1101/2023.03.05.531183.
Wang WW, Chen LY, Wozniak JM, Jadhav AM, Anderson H, Malone TE, et al. Targeted protein acetylation in cells using heterobifunctional molecules. J Am Chem Soc. 2021;143:16700–8.
Raina K, Forbes CD, Stronk R, Rappi JP, Eastman KJ, Gerritz SW, et al. Regulated induced proximity targeting chimeras (RIPTACs): a novel heterobifunctional small molecule therapeutic strategy for killing cancer cells selectively. bioRxiv. 2023. https://doi.org/10.1101/2023.01.01.522436.
Yu X, Eastman KJ, Raina K, Jones KM, Forbes CD, Hundt A, et al. Prostate cancer RIPTAC™ therapeutics demonstrate activity in preclinical models of Enzalutamide-resistant prostate cancer. In: Proceedings of the American Association for Cancer Research. 2023;83(7_Supplement):1629. https://doi.org/10.1158/1538-7445.AM2023-1629.
Raina K, Eastman KJ, Yu X, Forbes CD, Jones KM, Mousseau JJ, et al. An oral androgen receptor RIPTAC for prostate cancer. J Clin Oncol. 2023;41:184
Gerritz SW, Kayser-Bricker KJ, Neklesa T, Puleo DE, Mousseau JJ, Zaware N, et al., inventorsHeterobifunctional compounds and their use in treating disease. Patent WO/2023/059609. 2023.
Gerritz SW., Kayser-Bricker KJ., Eastman KJ., Neklesa T, Raina KS, inventorsHeterobifunctional compounds and their use in treating disease. Patent WO/2023/059583. 2023.
Xue G, Wang K, Zhou D, Zhong H, Pan Z. Light-induced protein degradation with photocaged PROTACs. J Am Chem Soc. 2019;141:18370–4.
Jin YH, Lu MC, Wang Y, Shan WX, Wang XY, You QD, et al. Azo-PROTAC: novel light-controlled small-molecule tool for protein knockdown. J Med Chem. 2020;63:4644–54.
Reynders M, Trauner D. Optical control of targeted protein degradation. Cell Chem Biol. 2021;28:969–86.
Naro Y, Darrah K, Deiters A. Optical control of small molecule-induced protein degradation. J Am Chem Soc. 2020;142:2193–7.
Banik SM, Pedram K, Wisnovsky S, Ahn G, Riley NM, Bertozzi CR. Lysosome-targeting chimaeras for degradation of extracellular proteins. Nature. 2020;584:291–7.
Ramadas B, Kumar Pain P, Manna D. LYTACs: an emerging tool for the degradation of non-cytosolic proteins. ChemMedChem. 2021;16:2951–3.
Mondal B, Dutta T, Padhy A, Das S, Sen Gupta S. Lysosome-targeting strategy using polypeptides and chimeric molecules. ACS Omega. 2022;7:5–16.
Hong D, Zhou B, Zhang B, Ren H, Zhu L, Zheng G, et al. Recent advances in the development of EGFR degraders: PROTACs and LYTACs. Eur J Med Chem. 2022;239:114533.
Ahn G, Banik SM, Miller CL, Riley NM, Cochran JR, Bertozzi CR. LYTACs that engage the asialoglycoprotein receptor for targeted protein degradation. Nat Chem Biol. 2021;17:937–46.
Wu Y, Lin B, Lu Y, Li L, Deng K, Zhang S, et al. Aptamer-LYTACs for targeted degradation of extracellular and membrane. Proteins Angew Chem Int Ed Engl. 2023;62:e202218106.
Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol. 2010;221:3–12.
Takahashi D, Moriyama J, Nakamura T, Miki E, Takahashi E, Sato A, et al. AUTACs: cargo-specific degraders using selective autophagy. Mol Cell. 2019;76:797–810 e10.
Pei J, Pan X, Wang A, Shuai W, Bu F, Tang P, et al. Developing potent LC3-targeting AUTAC tools for protein degradation with selective autophagy. Chem Commun. 2021;57:13194–7.
Ji CH, Kim HY, Lee MJ, Heo AJ, Park DY, Lim S, et al. The AUTOTAC chemical biology platform for targeted protein degradation via the autophagy-lysosome system. Nat Commun. 2022;13:904.
Li Z, Zhu C, Ding Y, Fei Y, Lu B. ATTEC: a potential new approach to target proteinopathies. Autophagy. 2020;16:185–7.
Buckley DL, Raina K, Darricarrere N, Hines J, Gustafson JL, Smith IE, et al. HaloPROTACS: use of small molecule PROTACs to induce degradation of HaloTag fusion proteins. ACS Chem Biol. 2015;10:1831–7.
Tovell H, Testa A, Maniaci C, Zhou H, Prescott AR, Macartney T, et al. Rapid and reversible knockdown of endogenously tagged endosomal proteins via an optimized HaloPROTAC degrader. ACS Chem Biol. 2019;14:882–92.
Bond AG, Craigon C, Chan KH, Testa A, Karapetsas A, Fasimoye R, et al. Development of BromoTag: a “bump-and-hole”-PROTAC system to induce potent, rapid, and selective degradation of tagged target proteins. J Med Chem. 2021;64:15477–502.
Simpson LM, Macartney TJ, Nardin A, Fulcher LJ, Roth S, Testa A, et al. Inducible degradation of target proteins through a tractable affinity-directed protein missile system. Cell Chem Biol. 2020;27:1164–80 e5.
Chen W, Younis MH, Zhao Z, Cai W. Recent biomedical advances enabled by HaloTag technology. Biocell. 2022;46:1789–801.
Grohmann C, Magtoto CM, Walker JR, Chua NK, Gabrielyan A, Hall M, et al. Development of NanoLuc-targeting protein degraders and a universal reporter system to benchmark tag-targeted degradation platforms. Nat Commun. 2022;13:2073.
Zhang H, Han Y, Yang Y, Lin F, Li K, Kong L, et al. Covalently engineered nanobody chimeras for targeted membrane protein degradation. J Am Chem Soc. 2021;143:16377–82.
Kim H, Park J, Kim JM. Targeted protein degradation to overcome resistance in cancer therapies: PROTAC and N-Degron pathway. Biomedicines. 2022;10:9.
Costales MG, Matsumoto Y, Velagapudi SP, Disney MD. Small molecule targeted recruitment of a nuclease to RNA. J Am Chem Soc. 2018;140:6741–4.
Gagliardi M, Ashizawa AT. The challenges and strategies of antisense oligonucleotide drug delivery. Biomedicines. 2021;9:4.
Velagapudi SP, Cameron MD, Haga CL, Rosenberg LH, Lafitte M, Duckett DR, et al. Design of a small molecule against an oncogenic noncoding RNA. Proc Natl Acad Sci USA. 2016;113:5898–903.
Meyer SM, Tanaka T, Zanon PRA, Baisden JT, Abegg D, Yang X, et al. DNA-encoded library screening to inform design of a ribonuclease targeting chimera (RiboTAC). J Am Chem Soc. 2022;144:21096–102.
Ishida T, Ciulli A. E3 ligase ligands for PROTACs: how they were found and how to discover new ones. SLAS Discov. 2021;26:484–502.
Ghidini A, Clery A, Halloy F, Allain FHT, Hall J. RNA-PROTACs: degraders of RNA-binding. Proteins Angew Chem Int Ed Engl. 2021;60:3163–9.
Eckstein F. Phosphorothioate oligodeoxynucleotides: what is their origin and what is unique about them? Antisense Nucleic Acid Drug Dev. 2000;10:117–21.
Egli M, Manoharan M. Re-engineering RNA molecules into therapeutic agents. Acc Chem Res. 2019;52:1036–47.
Samarasinghe KTG, Jaime-Figueroa S, Burgess M, Nalawansha DA, Dai K, Hu Z, et al. Targeted degradation of transcription factors by TRAFTACs: TRAnscription factor targeting chimeras. Cell Chem Biol. 2021;28:648–61 e5.
Samarasinghe KTG, An E, Genuth MA, Chu L, Holley SA, Crews CM. OligoTRAFTACs: a generalizable method for transcription factor degradation. RSC Chem Biol. 2022;3:1144–53.
Lim S, Khoo R, Peh KM, Teo J, Chang SC, Ng S, et al. bioPROTACs as versatile modulators of intracellular therapeutic targets including proliferating cell nuclear antigen (PCNA). Proc Natl Acad Sci USA. 2020;117:5791–800.
Zhao L, Ren TH, Wang DD. Clinical pharmacology considerations in biologics development. Acta Pharm Sin. 2012;33:1339–47.
Anselmo AC, Gokarn Y, Mitragotri S. Non-invasive delivery strategies for biologics. Nat Rev Drug Discov. 2019;18:19–40.
Krah S, Kolmar H, Becker S, Zielonka S. Engineering IgG-like bispecific antibodies-an overview. Antibodies. 2018;7:3.
Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192:1027–34.
Doroshow DB, Bhalla S, Beasley MB, Sholl LM, Kerr KM, Gnjatic S, et al. PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat Rev Clin Oncol. 2021;18:345–62.
Gadd MS, Testa A, Lucas X, Chan K-H, Chen W, Lamont DJ, et al. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat Chem Biol. 2017;13:514–21.
Riching KM, Mahan S, Corona CR, Mcdougall M, Vasta JD, Robers MB, et al. Quantitative live-cell kinetic degradation and mechanistic profiling of PROTAC mode of action. ACS Chem Biol. 2018;13:2758–70.
Roy MJ, Winkler S, Hughes SJ, Whitworth C, Galant M, Farnaby W, et al. SPR-measured dissociation kinetics of PROTAC ternary complexes influence target degradation rate. ACS Chem Biol. 2019;14:361–8.
Weiss DR, Bortolato A, Sun Y, Cai X, Lai C, Guo S, et al. On ternary complex stability in protein degradation: in silico molecular glue binding affinity calculations. J Chem Inf Model. 2023;63:2382–92.
Zorba A, Nguyen C, Xu Y, Starr J, Borzilleri K, Smith J, et al. Delineating the role of cooperativity in the design of potent PROTACs for BTK. Proc Natl Acad Sci USA. 2018;115:E7285–E92.
Nowak RP, Deangelo SL, Buckley D, He Z, Donovan KA, An J, et al. Plasticity in binding confers selectivity in ligand-induced protein degradation. Nat Chem Biol. 2018;14:706–14.
Drummond ML, Williams CI. In silico modeling of PROTAC-mediated ternary complexes: validation and application. J Chem Inf Model. 2019;59:1634–44.
Drummond ML, Henry A, Li H, Williams CI. Improved accuracy for modeling PROTAC-mediated ternary complex formation and targeted protein degradation via new in silico methodologies. J Chem Inf Model. 2020;60:5234–54.
Hawkins PC, Skillman AG, Warren GL, Ellingson BA, Stahl MT. Conformer generation with OMEGA: algorithm and validation using high quality structures from the Protein Databank and Cambridge Structural Database. J Chem Inf Model. 2010;50:572–84.
Bai N, Miller SA, Andrianov GV, Yates M, Kirubakaran P, Karanicolas J. Rationalizing PROTAC-mediated ternary complex formation using Rosetta. J Chem Inf Model. 2021;61:1368–82.
Bai N, Riching KM, Makaju A, Wu H, Acker TM, Ou SC, et al. Modeling the CRL4A ligase complex to predict target protein ubiquitination induced by cereblon-recruiting PROTACs. J Biol Chem. 2022;298:101653.
Zaidman D, Prilusky J, London N. PRosettaC: Rosetta based modeling of PROTAC mediated ternary complexes. J Chem Inf Model. 2020;60:4894–903.
Duhovny D, Nussinov R, Wolfson HJ. Efficient unbound docking of rigid molecules. Lect Notes Comput Sc. 2002;2452:185–200.
Rodriguez-Granillo A. Computational workflows for bifunctional degrader design. In: 3rd annual targeted protein degradation summit. October 13–15, 2020. Hanson Wade Group.
Dan Sindhikara MW, Gkeka P, Güssregen S, Tiwari G, Hessler G, Yapici E, et al. Automated design of macrocycles for therapeutic applications: from small molecules to peptides and proteins. J Med Chem. 2020;63:12100–15.
Eron SJ, Huang H, Agafonov RV, Fitzgerald ME, Patel J, Michael RE, et al. Structural characterization of degrader-induced ternary complexes using hydrogen-deuterium exchange mass spectrometry and computational modeling: implications for structure-based design. ACS Chem Biol. 2021;16:2228–43.
Dixon T, Macpherson D, Mostofian B, Dauzhenka T, Lotz S, Mcgee D, et al. Predicting the structural basis of targeted protein degradation by integrating molecular dynamics simulations with structural mass spectrometry. Nat Commun. 2022;13:5884.
Farnaby W, Koegl M, Roy MJ, Whitworth C, Diers E, Trainor N, et al. BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design. Nat Chem Biol. 2019;15:672–80.
Dragovich PS, Pillow TH, Blake RA, Sadowsky JD, Adaligil E, Adhikari P, et al. Antibody-mediated delivery of chimeric BRD4 degraders. part 2: improvement of in vitro antiproliferation activity and in vivo antitumor efficacy. J Med Chem. 2021;64:2576–607.
Kofink C, Trainor N, Mair B, Wohrle S, Wurm M, Mischerikow N, et al. A selective and orally bioavailable VHL-recruiting PROTAC achieves SMARCA2 degradation in vivo. Nat Commun. 2022;13:5969.
Schiemer J, Horst R, Meng Y, Montgomery JI, Xu Y, Feng X, et al. Snapshots and ensembles of BTK and cIAP1 protein degrader ternary complexes. Nat Chem Biol. 2021;17:152–60.
Chung CW, Dai H, Fernandez E, Tinworth CP, Churcher I, Cryan J, et al. Structural insights into PROTAC-mediated degradation of Bcl-xL. ACS Chem Biol. 2020;15:2316–23.
Yu X, Li D, Kottur J, Shen Y, Kim HS, Park KS, et al. A selective WDR5 degrader inhibits acute myeloid leukemia in patient-derived mouse models. Sci Transl Med. 2021;13:eabj1578.
Law RP, Nunes J, Chung CW, Bantscheff M, Buda K, Dai H, et al. Discovery and characterisation of highly cooperative FAK-degrading PROTACs. Angew Chem Int Ed Engl. 2021;60:23327–34.
Rosenberg SC, Shanahan F, Yamazoe S, Kschonsak M, Zeng YJ, Lee J, et al. Ternary complex dissociation kinetics contribute to mutant-selective EGFR degradation. Cell Chem Biol. 2023;30:175–87.
Smith BE, Wang SL, Jaime-Figueroa S, Harbin A, Wang J, Hamman BD, et al. Differential PROTAC substrate specificity dictated by orientation of recruited E3 ligase. Nat Commun. 2019;10:131.
Posternak G, Tang X, Maisonneuve P, Jin T, Lavoie H, Daou S, et al. Functional characterization of a PROTAC directed against BRAF mutant V600E. Nat Chem Biol. 2020;16:1170–8.
Zoppi V, Hughes SJ, Maniaci C, Testa A, Gmaschitz T, Wieshofer C, et al. Iterative design and optimization of initially inactive proteolysis targeting chimeras (PROTACs) identify VZ185 as a potent, fast, and selective von Hippel-Lindau (VHL) based dual degrader probe of BRD9 and BRD7. J Med Chem. 2019;62:699–726.
Heim C, Pliatsika D, Mousavizadeh F, Bar K, Hernandez Alvarez B, Giannis A, et al. De-novo design of cereblon (CRBN) effectors guided by natural hydrolysis products of thalidomide derivatives. J Med Chem. 2019;62:6615–29.
Kaneshige A, Bai L, Wang M, Mceachern D, Meagher JL, Xu R, et al. A selective small-molecule STAT5 PROTAC degrader capable of achieving tumor regression in vivo. Nat Chem Biol. 2023;19:703–11.
Hanzl A, Casement R, Imrichova H, Hughes SJ, Barone E, Testa A, et al. Functional E3 ligase hotspots and resistance mechanisms to small-molecule degraders. Nat Chem Biol. 2023;19:323–33.
Vieux EF, Agafonov RV, Emerson L, Isasa M, Deibler RW, Simard JR, et al. A method for determining the kinetics of small-molecule-induced ubiquitination. SLAS Discov. 2021;26:547–59.
Riching KM, Caine EA, Urh M, Daniels DL. The importance of cellular degradation kinetics for understanding mechanisms in targeted protein degradation. Chem Soc Rev. 2022;51:6210–21.
Bartlett DW, Gilbert AM. A kinetic proofreading model for bispecific protein degraders. J Pharmacokinet Pharmacodyn. 2021;48:149–63.
Mahan SD, Riching KM, Urh M, Daniels DL. Kinetic detection of E3:PROTAC: target ternary complexes using NanoBRET technology in live cells. Methods Mol Biol. 2021;2365:151–71.
Riching KM, Vasta JD, Hughes SJ, Zoppi V, Maniaci C, Testa A, et al. Translating PROTAC chemical series optimization into functional outcomes underlying BRD7 and BRD9 protein degradation. Curr Res Chem Biol. 2021;1:100009.
Huang HT, Dobrovolsky D, Paulk J, Yang G, Weisberg EL, Doctor ZM, et al. A chemoproteomic approach to query the degradable kinome using a multi-kinase degrader. Cell Chem Biol. 2018;25:88–99 e6.
Scholes NS, Mayor-Ruiz C, Winter GE. Identification and selectivity profiling of small-molecule degraders via multi-omics approaches. Cell Chem Biol. 2021;28:1048–60.
Jiang F, Wei Q, Li H, Li H, Cui Y, Ma Y, et al. Discovery of novel small molecule induced selective degradation of the bromodomain and extra-terminal (BET) bromodomain protein BRD4 and BRD2 with cellular potencies. Bioorg Med Chem. 2020;28:115181.
Park D, Izaguirre J, Coffey R, Xu H. Modeling the effect of cooperativity in ternary complex formation and targeted protein degradation mediated by heterobifunctional degraders. ACS Bio Med Chem Au. 2023;3:74–86.
Weng G, Cai X, Cao D, Du H, Shen C, Deng Y, et al. PROTAC-DB 2.0: an updated database of PROTACs. Nucleic Acids Res. 2023;51:D1367–D72.
Weng G, Shen C, Cao D, Gao J, Dong X, He Q, et al. PROTAC-DB: an online database of PROTACs. Nucleic Acids Res. 2021;49:D1381–D7.
Schneider M, Radoux CJ, Hercules A, Ochoa D, Dunham I, Zalmas LP, et al. The PROTACtable genome. Nat Rev Drug Discov. 2021;20:789–97.
Han B. A suite of mathematical solutions to describe ternary complex formation and their application to targeted protein degradation by heterobifunctional ligands. J Biol Chem. 2020;295:15280–91.
Chaudhry C. A mathematical model for covalent proteolysis targeting chimeras: thermodynamics and kinetics underlying catalytic efficiency. chemrxiv. https://doi.org/10.26434/chemrxiv-2021-vw4qb. online ahead of print.
Zheng S, Tan Y, Wang Z, Li C, Zhang Z, Sang X, et al. Accelerated rational PROTAC design via deep learning and molecular simulations. Nat Mach Intell. 2022;4:739–48.
Imrie F, Bradley AR, Van Der Schaar M, Deane CM. Deep generative models for 3D linker design. J Chem Inf Model. 2020;60:1983–95.
Li F, Hu Q, Zhang X, Sun R, Liu Z, Wu S, et al. DeepPROTACs is a deep learning-based targeted degradation predictor for PROTACs. Nat Commun. 2022;13:7133.
Atilaw Y, Poongavanam V, Svensson Nilsson C, Nguyen D, Giese A, Meibom D, et al. Solution conformations shed light on PROTAC cell permeability. ACS Med Chem Lett. 2021;12:107–14.
Weerakoon D, Carbajo RJ, De Maria L, Tyrchan C, Zhao H. Impact of PROTAC linker plasticity on the solution conformations and dissociation of the ternary complex. J Chem Inf Model. 2022;62:340–9.
Volak LP, Duevel HM, Humphreys S, Nettleton D, Phipps C, Pike A, et al. Industry perspective on the pharmacokinetic and ADME characterization of heterobifunctional protein degraders. Drug Metab Dispos. 2023. https://doi.org/10.1124/dmd.122.001154. Online ahead of print.
Edmondson SD, Yang B, Fallan C. Proteolysis targeting chimeras (PROTACs) in ‘beyond rule-of-five’ chemical space: recent progress and future challenges. Bioorg Med Chem Lett. 2019;29:1555–64.
Moreau K, Coen M, Zhang AX, Pachl F, Castaldi MP, Dahl G, et al. Proteolysis-targeting chimeras in drug development: a safety perspective. Br J Pharm. 2020;177:1709–18.
Acknowledgements
The authors would like to thank Christoph Zapf and Jodi Muckelbauer for careful review of the manuscript and Isha Verma for helpful discussions and review of figures.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors are currently employed by Bristol Myers Squibb and are shareholders of Bristol Myers Squibb.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hollingsworth, S., Johnson, S., Khakbaz, P. et al. The rise of targeting chimeras (TACs): next-generation medicines that preempt cellular events. Med Chem Res 32, 1294–1314 (2023). https://doi.org/10.1007/s00044-023-03104-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-023-03104-z