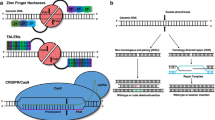
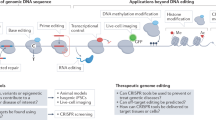
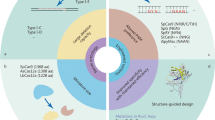
Abstract
Metabolic and cardiovascular diseases are world-concerning pathologies that affect an important percentage of the population. Nowadays, advances in the genetic background of these diseases allow new approaches to models and therapies, as well as different gene edition trials. Furthermore, technological improvements in gene editing go along with the development of new online and biocomputational tools that provide us alternative ways to explore pathologies. In this chapter, historical gene editing methods are discussed but focusing on CRISPR-Cas system in detail and also online resources available to perform these types of experiments. Here, the different strategies for gene editing and their online tools are gathered, putting the light on its application in the study and treatment of cardiovascular and metabolic diseases.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Pierpont ME, Brueckner M, Chung WK, Garg V, Lacro RV, McGuire AL, Mital S, Priest JR, Pu WT, Roberts A, Ware SM, Gelb BD, Russell MW, American Heart Association Council on Cardiovascular Disease in the Young; Council on Cardiovascular and Stroke Nursing; and Council on Genomic and Precision Medicine (2018) Genetic basis for congenital heart disease: revisited: A scientific statement from the American Heart Association. Circulation 138(21):e653–e711
Marian AJ, Asatryan B, Wehrens XHT (2020) Genetic basis and molecular biology of cardiac arrhythmias in cardiomyopathies. Cardiovasc Res 116(9):1600–1619
Tobita T, Nomura S, Fujita T, Morita H, Asano Y, Onoue K, Ito M, Imai Y, Suzuki A, Ko T, Satoh M, Fujita K, Naito AT, Furutani Y, Toko H, Harada M, Amiya E, Hatano M, Takimoto E, Shiga T, Nakanishi T, Sakata Y, Ono M, Saito Y, Takashima S, Hagiwara N, Aburatani H, Komuro I (2018) Genetic basis of cardiomyopathy and the genotypes involved in prognosis and left ventricular reverse remodeling. Sci Rep 8(1):1998
Krittanawong C, Sun T, Herzog E (2017) Big data and genome editing technology: a new paradigm of cardiovascular genomics. Curr Cardiol Rev 13(4):301–304
Takeda N, Komuro I (2019) Genetic basis of hereditary thoracic aortic aneurysms and dissections. J Cardiol 74(2):136–143
Maule G, Arosio D, Cereseto A (2020) Gene therapy for cystic fibrosis: progress and challenges of genome editing. Int J Mol Sci 21(11):3903
Kreindler JL (2010) Cystic fibrosis: exploiting its genetic basis in the hunt for new therapies. J Clin Pharm Ther 125(2):219–229
Wan T, Ping Y (2020) Delivery of genome-editing biomacromolecules for treatment of lung genetic disorders. Adv Drug Deliv Rev 168:196–216
Pividori M, Schoettler N, Nicolae DL, Ober C, Im HK (2019) Shared and distinct genetic risk factors for childhood-onset and adult-onset asthma: genome-wide and transcriptome-wide studies. Lancet Respir Med 7(6):509–522
Kim D, Lee YS, Kim DH, Bae SC (2020) Lung cancer staging and associated genetic and epigenetic events. Mol Cells 43(1):1–9
Xu X, Ng B, Sim B, Radulescu CI, Yusof NABM, Goh WI, Lin S, Lim JSY, Cha Y, Kusko R (2020) pS421 huntingtin modulates mitochondrial phenotypes and confers neuroprotection in an HD hiPSC model. Cell Death Dis 11(9):1–12
Ghosh R, Tabrizi SJ (2018) Clinical features of Huntington’s disease. Adv Exp Med Biol 1049:1–28
Ghosh R, Tabrizi SJ (2018) Huntington disease. Handb Clin Neurol 147:255–278
Mencacci NE, Carecchio M (2016) Recent advances in genetics of chorea. Curr Opin Neurol 29(4):486–495
Vermersch E, Jouve C, Hulot JS (2020) CRISPR/Cas9 gene-editing strategies in cardiovascular cells. Cardiovasc Res 116(5):894–907
Chow MT, Chang RYK, Chan H-K (2020) Inhalation delivery technology for genome-editing of respiratory diseases. Adv Drug Deliv Rev 168:217–228
Xu X, Wan T, Xin H, Li D, Pan H, Wu J, Ping Y (2019) Delivery of CRISPR/Cas9 for therapeutic genome editing. J Gene Med 21(7):e3107
Carroll D (2017) Genome editing: past, present, and future. Yale J Biol Med 90(4):653–659
Kaur K, Tandon H, Gupta AK, Kumar M (2015) CrisprGE: a central hub of CRISPR/Cas-based genome editing. Database (Oxford) 2015:bav055
Sledzinski P, Nowaczyk M, Olejniczak M (2020) Computational tools and resources supporting CRISPR-Cas experiments. Cells 9(5):1288
Makarova KS, Wolf YI, Koonin EV (2013) Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res 41(8):4360–4377
Zhang F, Wen Y, Guo X (2014) CRISPR/Cas9 for genome editing: progress, implications and challenges. Hum Mol Genet 23(R1):R40–R46
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337(6096):816–821
Wu X, Kriz AJ, Sharp PA (2014) Target specificity of the CRISPR-Cas9 system. Quant Biol 2(2):59–70
Reddy P, Vilella F, Izpisua Belmonte JC, Simon C (2020) Use of customizable nucleases for gene editing and other novel applications. Genes (Basel) 11(9):976
Anzalone AV, Koblan LW, Liu DR (2020) Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol 38(7):824–844
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8(11):2281–2308
Miles LA, Garippa RJ, Poirier JT (2016) Design, execution, and analysis of pooled in vitro CRISPR/Cas9 screens. FEBS J 283(17):3170–3180
Moon SB, Kim DY, Ko JH, Kim YS (2019) Recent advances in the CRISPR genome editing tool set. Exp Mol Med 51(11):1–11
Harrison PT, Hart S (2018) A beginner’s guide to gene editing. Exp Physiol 103(4):439–448
Alkhnbashi OS, Meier T, Mitrofanov A, Backofen R, Voss B (2020) CRISPR-Cas bioinformatics. Methods 172:3–11
Liu Z, Dong H, Cui Y, Cong L, Zhang D (2020) Application of different types of CRISPR/Cas-based systems in bacteria. Microb Cell Fact 19(1):172
Nishida K, Kondo A (2021) CRISPR-derived genome editing technologies for metabolic engineering. Metab Eng 63:141–147
Liu T, Pan S, Li Y, Peng N, She Q (2018) Type III CRISPR-Cas system: introduction and its application for genetic manipulations. Curr Issues Mol Biol 26:1–14
Koonin EV, Makarova KS, Zhang F (2017) Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol 37:67–78
Li B, Zeng C, Dong Y (2018) Design and assessment of engineered CRISPR-Cpf1 and its use for genome editing. Nat Protoc 13(5):899–914
Burmistrz M, Krakowski K, Krawczyk-Balska A (2020) RNA-targeting CRISPR-Cas systems and their applications. Int J Mol Sci 21(3):1122
Manghwar H, Li B, Ding X, Hussain A, Lindsey K, Zhang X, Jin S (2020) CRISPR/Cas systems in genome editing: methodologies and tools for sgRNA design, off-target evaluation, and strategies to mitigate off-target effects. Adv Sci (Weinh) 7(6):1902312
Zhang JH, Adikaram P, Pandey M, Genis A, Simonds WF (2016) Optimization of genome editing through CRISPR-Cas9 engineering. Bioengineered 7(3):166–174
Ma Y, Zhang L, Huang X (2014) Genome modification by CRISPR/Cas9. FEBS J 281(23):5186–5193
Fung K (2019) Detection of sickle cell disease-associated single nucleotide polymorphism using a graphene field effect transistor. Keck Science Department of Claremont McKenna, Pitzer, and Scripps Colleges. CMC Senior Theses. 2262. https://scholarship.claremont.edu/cmc_theses/2262
Schaefer KA, Wu WH, Colgan DF, Tsang SH, Bassuk AG, Mahajan VB (2017) Unexpected mutations after CRISPR-Cas9 editing in vivo. Nat Methods 14(6):547–548
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339(6121):819–823
Wang J, Zhang X, Cheng L, Luo Y (2020) An overview and metanalysis of machine and deep learning-based CRISPR gRNA design tools. RNA Biol 17(1):13–22
Wienert B, Wyman SK, Richardson CD, Yeh CD, Akcakaya P, Porritt MJ, Morlock M, Vu JT, Kazane KR, Watry HL, Judge LM, Conklin BR, Maresca M, Corn JE (2019) Unbiased detection of CRISPR off-targets in vivo using DISCOVER-Seq. Science 364(6437):286–289
Naito Y, Hino K, Bono H, Ui-Tei K (2015) CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31(7):1120–1123
Lee J, Jung MH, Jeong E, Lee JK (2019) Using Sniper-Cas9 to minimize off-target effects of CRISPR-Cas9 without the loss of on-target activity via directed evolution. J Vis Exp 144:doi:10.3791/59202
Yang L, Yang JL, Byrne S, Pan J, Church GM (2014) CRISPR/Cas9-directed genome editing of cultured cells. Curr Protoc Mol Biol 107:31.1.1–31.117
Guilinger JP, Thompson DB, Liu DR (2014) Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol 32(6):577–582
Hanna RE, Doench JG (2020) Design and analysis of CRISPR-Cas experiments. Nat Biotechnol 38(7):813–823
Kleinstiver BP, Sousa AA, Walton RT, Tak YE, Hsu JY, Clement K, Welch MM, Horng JE, Malagon-Lopez J, Scarfo I, Maus MV, Pinello L, Aryee MJ, Joung JK (2019) Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat Biotechnol 37(3):276–282
Xu X, Duan D, Chen SJ (2017) CRISPR-Cas9 cleavage efficiency correlates strongly with target-sgRNA folding stability: from physical mechanism to off-target assessment. Sci Rep 7(1):143
Filippova J, Matveeva A, Zhuravlev E, Stepanov G (2019) Guide RNA modification as a way to improve CRISPR/Cas9-based genome-editing systems. Biochimie 167:49–60
Moon SB, Kim DY, Ko JH, Kim JS, Kim YS (2019) Improving CRISPR genome editing by engineering guide RNAs. Trends Biotechnol 37(8):870–881
Mir A, Alterman JF, Hassler MR, Debacker AJ, Hudgens E, Echeverria D, Brodsky MH, Khvorova A, Watts JK, Sontheimer EJ (2018) Heavily and fully modified RNAs guide efficient SpyCas9-mediated genome editing. Nat Commun 9(1):2641
Zhang JP, Li XL, Neises A, Chen W, Hu LP, Ji GZ, Yu JY, Xu J, Yuan WP, Cheng T, Zhang XB (2016) Different effects of sgRNA length on CRISPR-mediated gene knockout efficiency. Sci Rep 6:28566
Ryu SM, Koo T, Kim K, Lim K, Baek G, Kim ST, Kim HS, Kim DE, Lee H, Chung E, Kim JS (2018) Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat Biotechnol 36(6):536–539
Lee K, Mackley VA, Rao A, Chong AT, Dewitt MA, Corn JE, Murthy N (2017) Synthetically modified guide RNA and donor DNA are a versatile platform for CRISPR-Cas9 engineering. Elife 6:e25312
Cui Y, Xu J, Cheng M, Liao X, Peng S (2018) Review of CRISPR/Cas9 sgRNA design tools. Interdiscip Sci 10(2):455–465
Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, Root DE (2014) Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol 32(12):1262–1267
Xu H, Xiao T, Chen CH, Li W, Meyer CA, Wu Q, Wu D, Cong L, Zhang F, Liu JS, Brown M, Liu XS (2015) Sequence determinants of improved CRISPR sgRNA design. Genome Res 25(8):1147–1157
Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, Virgin HW, Listgarten J, Root DE (2016) Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34(2):184–191
Pallares Masmitja M, Knodlseder N, Guell M (2019) CRISPR-gRNA Design. Methods Mol Biol 1961:3–11
Seki A, Rutz S (2018) Optimized RNP transfection for highly efficient CRISPR/Cas9-mediated gene knockout in primary T cells. J Exp Med 215(3):985–997
Laughery MF, Hunter T, Brown A, Hoopes J, Ostbye T, Shumaker T, Wyrick JJ (2015) New vectors for simple and streamlined CRISPR-Cas9 genome editing in Saccharomyces cerevisiae. Yeast 32(12):711–720
DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM (2013) Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res 41(7):4336–4343
Farooq R, Hussain K, Tariq M, Farooq A, Mustafa M (2020) CRISPR/Cas9: targeted genome editing for the treatment of hereditary hearing loss. J Appl Genet 61(1):51–65
Zhang J, Chen L, Zhang J, Wang Y (2019) Drug inducible CRISPR/Cas Systems. Comput Struct Biotechnol J 17:1171–1177
Vad-Nielsen J, Lin L, Bolund L, Nielsen AL, Luo Y (2016) Golden gate assembly of CRISPR gRNA expression array for simultaneously targeting multiple genes. Cell Mol Life Sci 73(22):4315–4325
Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, Xie Y, Shen R, Chen S, Wang Z, Chen Y, Guo J, Chen L, Zhao X, Dong Z, Liu YG (2015) A Robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8(8):1274–1284
Li Y, Li S, Wang J, Liu G (2019) CRISPR/Cas systems towards next-generation biosensing. Trends Biotechnol 37(7):730–743
Wolter F, Puchta H (2018) Application of CRISPR/Cas to understand Cis- and trans-regulatory elements in plants. Methods Mol Biol 1830:23–40
Sanjana NE, Shalem O, Zhang F (2014) Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11(8):783–784
Bhatia A, Sra J, Akhtar M (2016) Preexcitation syndromes. Curr Prob Cardiol 41(3):99–137
Xie C, Zhang YP, Song L, Luo J, Qi W, Hu J, Lu D, Yang Z, Zhang J, Xiao J, Zhou B, Du JL, Jing N, Liu Y, Wang Y, Li BL, Song BL, Yan Y (2016) Genome editing with CRISPR/Cas9 in postnatal mice corrects PRKAG2 cardiac syndrome. Cell Res 26(10):1099–1111
Stehle R, Iorga B (2010) Kinetics of cardiac sarcomeric processes and rate-limiting steps in contraction and relaxation. J Mol Cell Cardiol 48(5):843–850
Mosqueira D, Mannhardt I, Bhagwan JR, Lis-Slimak K, Katili P, Scott E, Hassan M, Prondzynski M, Harmer SC, Tinker A, Smith JGW, Carrier L, Williams PM, Gaffney D, Eschenhagen T, Hansen A, Denning C (2018) CRISPR/Cas9 editing in human pluripotent stem cell-cardiomyocytes highlights arrhythmias, hypocontractility, and energy depletion as potential therapeutic targets for hypertrophic cardiomyopathy. Eur Heart J 39(43):3879–3892
Dick E, Matsa E, Young LE, Darling D, Denning C (2011) Faster generation of hiPSCs by coupling high-titer lentivirus and column-based positive selection. Nat Protoc 6(6):701–714
Karakikes I, Stillitano F, Nonnenmacher M, Tzimas C, Sanoudou D, Termglinchan V, Kong CW, Rushing S, Hansen J, Ceholski D, Kolokathis F, Kremastinos D, Katoulis A, Ren L, Cohen N, Gho J, Tsiapras D, Vink A, Wu JC, Asselbergs FW, Li RA, Hulot JS, Kranias EG, Hajjar RJ (2015) Correction of human phospholamban R14del mutation associated with cardiomyopathy using targeted nucleases and combination therapy. Nat Commun 6:6955
Chen Y, Lu W, Gao N, Long Y, Shao Y, Liu M, Chen H, Ye S, Ma X, Liu M, Li D (2017) Generation of obese rat model by transcription activator-like effector nucleases targeting the leptin receptor gene. Sci China Life Sci 60(2):152–157
Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B (2011) Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol 29(8):699–700
Acknowledgments
Julio Plaza-Diaz is part of the “UGR Plan Propio de Investigación 2016” and the “Excellence actions: Unit of Excellence on Exercise and Health (UCEES), University of Granada.” Julio Plaza-Diaz is supported by a grant awarded to postdoctoral researchers at foreign universities and research centers from the “Fundación Ramón Areces,” Madrid, Spain.
Competing Financial Interests
The authors declare no competing financial interests.
Funding
This research received no external funding.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Carrillo-Rodriguez, P. et al. (2023). Online Databases of Genome Editing in Cardiovascular and Metabolic Diseases. In: Xiao, J. (eds) Genome Editing in Cardiovascular and Metabolic Diseases. Advances in Experimental Medicine and Biology, vol 1396. Springer, Singapore. https://doi.org/10.1007/978-981-19-5642-3_2
Download citation
DOI: https://doi.org/10.1007/978-981-19-5642-3_2
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-19-5641-6
Online ISBN: 978-981-19-5642-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)