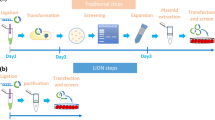
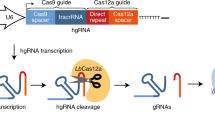
Abstract
The engineered CRISPR/Cas9 technology has developed as the most efficient and broadly used genome editing tool. However, simultaneously targeting multiple genes (or genomic loci) in the same individual cells using CRISPR/Cas9 remain one technical challenge. In this article, we have developed a Golden Gate Assembly method for the generation of CRISPR gRNA expression arrays, thus enabling simultaneous gene targeting. Using this method, the generation of CRISPR gRNA expression array can be accomplished in 2 weeks, and contains up to 30 gRNA expression cassettes. We demonstrated in the study that simultaneously targeting 10 genomic loci or simultaneously inhibition of multiple endogenous genes could be achieved using the multiplexed gRNA expression array vector in human cells. The complete set of plasmids is available through the non-profit plasmid repository Addgene.






Similar content being viewed by others
Notes
All gRNA expression cassettes in our method are driven by the U6 promoter.
It is very important to treat the reaction with plasmid safe DNA nuclease when assembling more than five gRNA expression cassettes. This step is to avoid the recombination of the linearized DNA fragments following transformation.
References
Cong L, Ran FA, Cox D et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823
Jinek M, Chylinski K, Fonfara I et al (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821
Wan H, Feng C, Teng F et al (2015) One-step generation of p53 gene biallelic mutant Cynomolgus monkey via the CRISPR/Cas system. Cell Res 25:258–261
Mali P, Yang L, Esvelt KM et al (2013) RNA-Guided human genome engineering via Cas9. Science 339:823–826
Cheng AW, Wang H, Yang H et al (2013) Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res 23:1163–1171
Gilbert LA, Larson MH, Morsut L et al (2013) CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154:442–451
Hilton IB, D’Ippolito AM, Vockley CM et al (2015) Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 33:510–517
Chen B, Gilbert LA, Cimini BA et al (2013) Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155:1479–1491
Engler C, Kandzia R, Marillonnet S (2008) A one pot, one step, precision cloning method with high throughput capability. PLoS ONE 3:e3647
Cermak T, Doyle EL, Christian M et al (2011) Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39:e82
Mandal PK, Ferreira LM, Collins R et al (2014) Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell 15:643–652
Zhou Y, Liu Y, Hussmann D et al (2016) Enhanced genome editing in mammalian cells with a modified dual-fluorescent surrogate system. Cell Mol Life Sci. doi:10.1007/s00018-015-2128-3
Qi LS, Larson MH, Gilbert LA et al (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152:1173–1183
Tu Z, Yang W, Yan S et al (2015) CRISPR/Cas9: a powerful genetic engineering tool for establishing large animal models of neurodegenerative diseases. Mol Neurodegener 10:35
Holm IE, Alstrup AK, Luo Y (2016) Genetically modified pig models for neurodegenerative disorders. J Pathol 238:267–287
Cooper DK, Ekser B, Ramsoondar J et al (2016) The role of genetically engineered pigs in xenotransplantation research. J Pathol 238:288–299
Kearns NA, Pham H, Tabak B et al (2015) Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods 12:401–403
Konermann S, Brigham MD, Trevino AE et al (2014) Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517:583–588. doi:10.1038/nature14136
Gilbert LA, Horlbeck MA, Adamson B et al (2014) Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159:647–661
Fujita T, Fujii H (2013) Efficient isolation of specific genomic regions and identification of associated proteins by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) using CRISPR. Biochem Biophys Res Commun 439:132–136
Ran FA, Hsu PD, Lin CY et al (2013) Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154:1380–1389
Slaymaker IM, Gao L, Zetsche B et al (2016) Rationally engineered Cas9 nucleases with improved specificity. Science 351:84–88
Kleinstiver BP, Pattanayak V, Prew MS et al (2016) High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529:490–495
Fu Y, Sander JD, Reyon D, et al (2014) Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 32(3):279–284. doi:10.1038/nbt.2808
Balboa D, Weltner J, Eurola S et al (2015) Conditionally stabilized dCas9 activator for controlling gene expression in human cell reprogramming and differentiation. Stem Cell Reports 5:448–459
Thomsen R, Solvsten CA, Linnet TE et al (2010) Analysis of qPCR data by converting exponentially related Ct values into linearly related X0 values. J Bioinform Comput Biol 8:885–900
Acknowledgments
JVN and ALN were supported by grants from The Lundbeck Foundation, Krista og Viggo Petersens Fond, Fabrikant Einar Willumsens Mindelegat, and Fonden til Lægevidenskabens Fremme. LL and LB were supported by the DREAM project from Lundbeck Foundation. YL was supported by grants from Danish Research Council for Independent Research, the Sapere Aude Young Research Talent prize to YL, the Lundbeck Foundation and the Innovation Fund Denmark (BrainStem).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Vad-Nielsen, J., Lin, L., Bolund, L. et al. Golden Gate Assembly of CRISPR gRNA expression array for simultaneously targeting multiple genes. Cell. Mol. Life Sci. 73, 4315–4325 (2016). https://doi.org/10.1007/s00018-016-2271-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-016-2271-5