Abstract
In a scenario of climate change and rapidly rising urban populations demanding processed foods, it is necessary to develop new wheat cultivars combining high yield potential, disease resistance, and stability for yield and processing quality, even under heat or drought stress conditions. Allelic variation for gluten proteins (glutenin subunits and gliadins) is a major determinant of differences in dough viscoelastic properties observed between cultivars of both bread wheat and durum wheat. Technical difficulties in allelic identification due to the complexity of the protein profile produced by each cultivar and the use of different nomenclature systems in different laboratories has historically interfered with information exchange between research groups, a situation exacerbated by the vast number of possible profiles found in different cultivars due to the multi-allelic nature of the principal loci encoding gluten proteins (Glu-1, Glu-2, Glu-3, Gli-1 and Gli-2). For the Glu-3 alleles, we have collaborated to unify criteria across laboratories and to compare four different methods of allelic identification (SDS-PAGE, 2-DE, MALDI-TOF-MS and PCR), and have shown that the four methods can be regarded as complementary techniques for allelic identification. We seek to continue addressing remaining analytical challenges, place the findings in the context of the Catalogue of Gene Symbols for Wheat, and, with unified criteria, initiate work to define better the relationship between specific gluten proteins and processing quality attributes. Therefore, we propose a new system to share materials through public gene banks in collaboration with the Catalogue, and the formation of a wider international group aimed at facilitating the resolution of the remaining problems in the field. We also propose to extend our collaboration by forming a wheat quality expert working group under the Wheat Initiative.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
Current Status of Glu-3 Allele Nomenclature
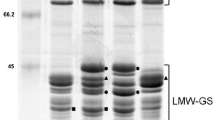
It has been shown that allelic variation for the high-molecular-weight glutenin subunits (HMW-GSs) and low-molecular-weight glutenin subunits (LMW-GSs) affects the properties of dough made with different wheat cultivars. LMW-GS composition in common wheat is one of the critical determinants of gluten properties (Branlard et al. 2001; Eagles et al. 2006; Gupta et al. 1994; Liu et al. 2005; Maruyama-Funatsuki et al. 2005). Gupta and Shepherd (1990) assigned the individual LMW-GSs to Glu-A3, Glu-B3 and Glu-D3 loci and selected standard cultivars that covered the allelic variation observed. However, subsequent use of Glu-3 nomenclature has not been consistent among laboratories, due to the complexity of the LMW-GSs, different separation methods and different standard cultivars used by researchers (Branlard et al. 2003; Ikeda et al. 2006; Singh et al. 1990). It is necessary to unify the various Glu-3 allelic nomenclature systems in use, to allow information to be shared regarding the effects of individual alleles on gluten properties and to be applied in breeding programs aimed at improving gluten properties. In previous studies, four laboratories plus an international institution shared cultivars and compared results. We confirmed that there were inconsistencies to identify Glu-3 alleles between laboratories due to differences of separation and identification methods (Ikeda et al. 2008). Using 2-DE analysis, we found new Glu-3 alleles among these materials (Ikeda et al. 2009). Combining SDS-PAGE, 2-DE, MALDI-TOF-MS and PCR analyses, we showed that a combination of methods was required to identify certain alleles, and that these methods would be especially useful when characterizing new alleles. We also recommended 30 cultivars as standards for the determination of LMW-GS alleles (Liu et al. 2010).
Sharing Materials
It is very important to share materials to unify nomenclature among research groups. However, it is not always possible to obtain cultivars representing Glu-3 alleles listed in Catalogue of Gene Symbols for Wheat (McIntosh et al. 2013). Therefore, we need to establish a system to share materials internationally. We propose to deposit cultivars representing particular alleles in public gene banks (e.g. Germplasm bank in CIMMYT, Genebank in VIR in Russia, NBRP in Japan, and GRIN in the USA). The registered alleles will be available publicly through these gene banks. New alleles can be evaluated by curators of the Catalogue and other researchers for registration in the catalogue. This system also helps to refine the catalogue (Fig. 31.1). At present CIMMYT Genebank performs seed multiplication of a Glu-3 common wheat master set.
Sharing Methods
It is also important to use common methods to identify Glu-3 alleles. For SDS-PAGE, Peña proposed the use of separation gels containing Tris buffer of pH 8.5 instead of pH 8.8 for better separation of LMW-GS bands (Ikeda et al. 2008; Peña et al. 2004). Lowering bis-acrylamide concentration and using larger size gels also helps better separation (Branlard et al. 2003). Further evaluation for creating a standard SDS-PAGE method is necessary. For PCR markers, as the number of known alleles increases, we need to reconfirm the usefulness of PCR markers to identify Glu-3 alleles. For example, a PCR primer set, which was developed to identify the Glu-B3i allele (Wang et al. 2009), identified the Glu-B3ad allele instead. It is necessary to select a standard PCR primer set to identify Glu-3 alleles.
Functional Analysis of Gluten Proteins
By sharing materials and methods among international research groups, it becomes possible to define better the relationship between specific gluten proteins and processing quality stability, even under heat or drought stress wheat growing conditions. We will set an international framework to evaluate allelic effects on quality attributes under various environmental conditions.
Unification with Durum Glu-3 Alleles
Durum Glu-3 alleles were classified independently of those of common wheat (Martinez et al. 2004; Nieto-Taladriz et al. 1997). In the Catalogue, the durum Glu-3 alleles were originally assigned separately and subsequently combined into one provisional list. Since tetraploid durum wheat shares common ancestral species with common wheat, we would expect some alleles to be identical to those of common wheat. We shared standard cultivars and studied Glu-3 alleles by SDS-PAGE, 2-DE and PCR. Some alleles seemed to share the same alleles with common wheat, but some were unique in durum wheat (data not shown). This means that durum allele might widen the genetic diversity of common wheat alleles, and vice versa. Further analysis is necessary to clarify durum Glu-3 alleles and produce a definitive list in the Catalogue for use by the wheat community. This is also important for Glu-1 alleles.
Gliadin Analysis
Gliadin consists of α/β/γ/ω-gliadins, which contain many proteins having a range of molecular weights and pI values. Variation in the gliadins also effects dough properties (Branlard and Dardevet 1994). Gliadins are also known to contain epitopes involved in wheat gluten related disorders (Sapone et al. 2012). Gliadin analysis was mainly carried out using A-PAGE. The analysis of gliadin proteins using SDS-PAGE allows the determination of the banding patterns associated with the close linkage existing between Gli-1 and Glu-3, and, therefore, this approach further contributes to the identification of specific Glu-3 LMW-GS in both common and durum wheat. With increasing genome sequence data availability, it is important to identify gliadins by proteomic techniques to clarify correspondence between gliadin proteins, the epitopes of allergen and coding genes (Juhasz et al. 2012).
Forming an International Gluten Research Group
To carry out these tasks, we propose to form an international gluten research group. Using the same materials under different environmental conditions makes it possible to evaluate the effects of Glu-3 alleles on dough properties in such conditions. From this collaboration, we can share advanced knowledge of gluten function for further study, accelerating the development of new cultivars maintaining good quality under climate change and responding to quality demands from industries and consumers (Fig. 31.2). It is also possible to use gluten protein alleles for cultivar identification to protect breeder’s rights.
Further Perspective
The field of gluten research overlaps other wheat quality related fields, e.g. allergy, nutrition and carbohydrates. Therefore, it is logical to attempt extending our collaboration to other researchers related to wheat quality. Currently we work to form a wheat quality expert working group under the Wheat Initiative (http://www.wheatinitiative.org/about/expert-working-groups). We would like to invite other colleagues related to wheat quality to join our collaboration.
References
Branlard G, Dardevet M (1994) A null Gli-D1 allele with a positive effect on bread wheat quality. J Cereal Sci 20:235–244
Branlard G, Dardevet M, Saccomano R et al (2001) Genetic diversity of wheat storage proteins and bread wheat quality. Euphytica 119:59–67
Branlard G, Dardevet M, Amiour N, Igrejas G (2003) Allelic diversity of HMW and LMW glutenin subunits and omega-gliadins in French bread wheat (Triticum aestivum L.). Genet Res Crop Evol 50:669–679
Eagles HA, Cane K, Eastwood RF et al (2006) Contributions of glutenin and puroindoline genes to grain quality traits in southern Australian wheat breeding programs. Aust J Agric Res 57:179–186
Gupta RB, Shepherd KW (1990) Two-step one-dimensional SDS-PAGE analysis of LMW subunits of glutelin. 1. Variation and genetic control of the subunits in hexaploid wheats. Theor Appl Genet 80:65–74
Gupta RB, Paul JG, Cornish GB et al (1994) Allelic variation at glutenin subunit and gliadin loci, Glu-1, Glu-3 and Gli-1, of common wheat. I. Its additive and interaction effects on dough properties. J Cereal Sci 19:9–17
Ikeda TM, Araki E, Fujita Y, Yano H (2006) Characterization of low-molecular-weight glutenin subunit genes and their protein products in common wheats. Theor Appl Genet 112:327–334
Ikeda TM, Branlard G, Peña RJ, et al (2008) International collaboration for unifying Glu-3 nomenclature system in common wheats. In: Appels R, Eastwood R, Lagudah E, et al (eds) The 11th international wheat genetics symposium, Sydney University Press, Brisbane
Ikeda TM, Branlard G, Peña RJ, et al (2009) Characterization of new Glu-3 alleles in bread wheat. In: INRA (ed) 10th international gluten workshop, INRA, Clermont-Ferrand, pp 180–184
Juhasz A, Gell G, Bekes F, Balazs E (2012) The epitopes in wheat proteins for defining toxic units relevant to human health. Funct Integr Genomics 12:585–598
Liu L, He Z, Yan J et al (2005) Allelic variation at the Glu-1 and Glu-3 loci, presence of the 1B.1R translocation, and their effects on mixographic properties in Chinese bread wheats. Euphytica 142:197–204
Liu L, Ikeda TM, Branlard G et al (2010) Comparison of low molecular weight glutenin subunits identified by SDS-PAGE, 2-DE. MALDI-TOF-MS and PCR in common wheat. BMC Plant Biol 10:124
Martinez MC, Ruiz M, Carrillo JM (2004) New B low Mr glutenin subunit alleles at the Glu-A3, Glu-B2 and Glu-B3 loci and their relationship with gluten strength in durum wheat. J Cereal Sci 40:101–107
Maruyama-Funatsuki W, Takata K, Funatsuki H et al (2005) Identification and characterization of a novel LMW-s glutenin gene of a Canadian Western extra-strong wheat. J Cereal Sci 41:47–57
McIntosh RA, Yamazaki Y, Dubcovsky J, et al (2013) Catalogue of gene symbols for wheat. In: 12th international wheat genetics symposium, Yokohama
Nieto-Taladriz MT, Ruiz M, Martínez MC et al (1997) Variation and classification of B low-molecular-weight glutenin subunit alleles in durum wheat. Theor Appl Genet 95:1155–1160
Peña RJ, Gonzalez-Santoyo H, Cervantes F (2004) Relationship between some Glu-D1/Glu-B3 allelic combinations and bread-making quality related parameters commonly used in wheat breeding. In: Lafiandra D, Masci S, D’Ovidio R (eds) The gluten proteins. Royal Society of Chemistry, Cambridge, pp 156–157b
Sapone A, Bai JC, Ciacci C et al (2012) Spectrum of gluten-related disorders: consensus on new nomenclature and classification. BMC Med 10:13
Singh NK, Shepherd KW, Cornish GB (1990) A simplified SDS-PAGE procedure for separating LMW subunits of glutenin. J Cereal Sci 14:203–208
Wang L, Zhao X, He Z et al (2009) Characterization of low-molecular-weight glutenin subunit Glu-B3 genes and development of STS markers in common wheat (Triticum aestivum L.). Theor Appl Genet 118:525–539
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is distributed under the terms of the Creative Commons Attribution Noncommercial License, which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Copyright information
© 2015 The Author(s)
About this paper
Cite this paper
Ikeda, T.M. et al. (2015). Proposal of International Gluten Research Group. In: Ogihara, Y., Takumi, S., Handa, H. (eds) Advances in Wheat Genetics: From Genome to Field. Springer, Tokyo. https://doi.org/10.1007/978-4-431-55675-6_31
Download citation
DOI: https://doi.org/10.1007/978-4-431-55675-6_31
Publisher Name: Springer, Tokyo
Print ISBN: 978-4-431-55674-9
Online ISBN: 978-4-431-55675-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)